6VOP
 
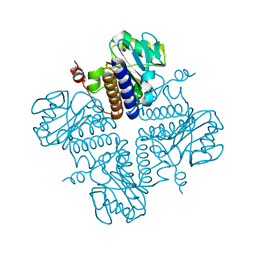 | | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Escherichia coli | | 分子名称: | Aldolase | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-01-31 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Escherichia coli
To Be Published
|
|
6W37
 
 | |
3M34
 
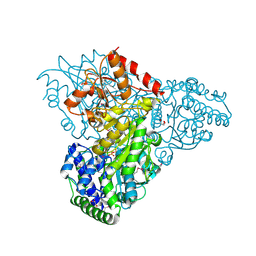 | | Crystal structure of transketolase in complex with thiamin diphosphate and calcium ion | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | 著者 | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-03-08 | | 公開日 | 2010-04-28 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of transketolase in complex with thiamin diphosphate and calcium ion
To be Published
|
|
4MBO
 
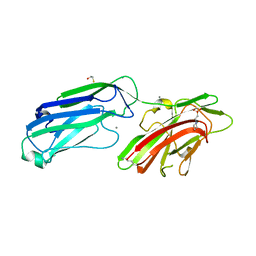 | | 1.65 Angstrom Crystal Structure of Serine-rich Repeat Adhesion Glycoprotein (Srr1) from Streptococcus agalactiae | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-08-19 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
3IV8
 
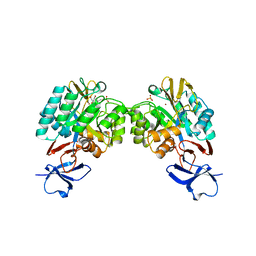 | | N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae complexed with fructose 6-phosphate | | 分子名称: | 6-O-phosphono-beta-D-fructofuranose, N-acetylglucosamine-6-phosphate deacetylase, NICKEL (II) ION, ... | | 著者 | Osipiuk, J, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-08-31 | | 公開日 | 2009-09-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | X-ray crystal structure of N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae complexed with fructose 6-phosphate.
To be Published
|
|
6W1W
 
 | | Crystal Structure of Motility Associated Killing Factor B from Vibrio cholerae | | 分子名称: | 1,2-ETHANEDIOL, motility-associated killing factor MakB | | 著者 | Kim, Y, Welk, L, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-04 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
5UXA
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphB from Escherichia coli | | 分子名称: | CALCIUM ION, Macrolide 2'-phosphotransferase II | | 著者 | Stogios, P.J, Evdokimova, E, Egorova, O, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-22 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
6W02
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | 著者 | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-28 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
5UXB
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium, apoenzyme | | 分子名称: | CHLORIDE ION, Macrolide 2'-phosphotransferase MphH | | 著者 | Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-22 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.794 Å) | | 主引用文献 | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5VKW
 
 | | Crystal structure of adenylosuccinate lyase ADE13 from Candida albicans | | 分子名称: | Adenylosuccinate lyase, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-04-24 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Crystal structure of adenylosuccinate lyase ADE13 from Candida albicans
To Be Published
|
|
6V98
 
 | | Crystal structure of Type VI secretion system effector, TseH (VCA0285) | | 分子名称: | CALCIUM ION, Cysteine hydrolase | | 著者 | Watanabe, N, Hersch, S.J, Dong, T.G, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Envelope stress responses defend against type six secretion system attacks independently of immunity proteins.
Nat Microbiol, 5, 2020
|
|
7JM2
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin | | 分子名称: | APRAMYCIN, Aminocyclitol acetyltransferase ApmA, CHLORIDE ION | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin
To Be Published
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | 著者 | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-18 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
3N2A
 
 | | Crystal structure of bifunctional folylpolyglutamate synthase/dihydrofolate synthase from Yersinia pestis CO92 | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bifunctional folylpolyglutamate synthase/dihydrofolate synthase, ... | | 著者 | Nocek, B, Maltseva, N, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-05-17 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of bifunctional folylpolyglutamate synthase/dihydrofolate synthase from Yersinia pestis CO92
TO BE PUBLISHED
|
|
6W63
 
 | |
6V82
 
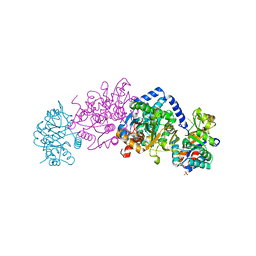 | | Crystal structure of tryptophan synthase from Chlamydia trachomatis D/UW-3/CX | | 分子名称: | SULFATE ION, Tryptophan synthase alpha chain, Tryptophan synthase beta chain, ... | | 著者 | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-10 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.424 Å) | | 主引用文献 | Catalytically impaired TrpA subunit of tryptophan synthase from Chlamydia trachomatis is an allosteric regulator of TrpB.
Protein Sci., 30, 2021
|
|
4MBR
 
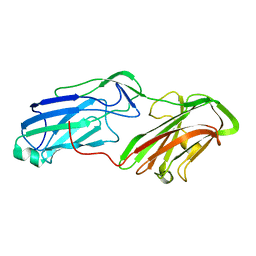 | | 3.65 Angstrom Crystal Structure of Serine-rich Repeat Protein (Srr2) from Streptococcus agalactiae | | 分子名称: | Serine-rich repeat protein 2 | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-08-19 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
6WCF
 
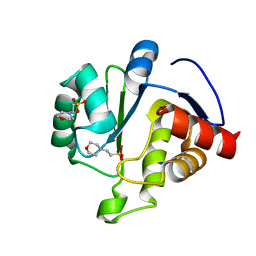 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural protein 3 | | 著者 | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-30 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.065 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
5VYV
 
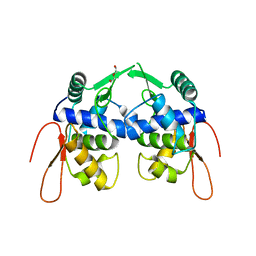 | | Crystal structure of a protein of unknown function YceH/ECK1052 involved in membrane biogenesis from Escherichia coli | | 分子名称: | DI(HYDROXYETHYL)ETHER, UPF0502 protein YceH | | 著者 | Stogios, P.J, Skarina, T, McChesney, C, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-05-26 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Crystal structure of a protein of unknown function YceH/ECK1052 involved in membrane biogenesis from Escherichia coli
To Be Published
|
|
6WDT
 
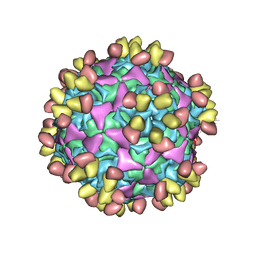 | | Enterovirus D68 in complex with human monoclonal antibody EV68-228 | | 分子名称: | EV68-228 heavy chain, EV68-228 light chain, viral protein 1, ... | | 著者 | Fu, J, Vogt, M.R, Klose, T, Crowe, J.E, Rossmann, M.G, Kuhn, R.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-01 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Human antibodies neutralize enterovirus D68 and protect against infection and paralytic disease.
Sci Immunol, 5, 2020
|
|
6WJT
 
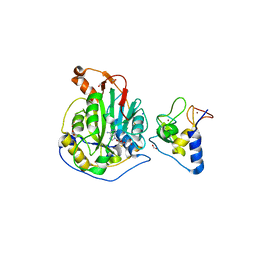 | | 2.0 Angstrom Resolution Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 in Complex with S-Adenosyl-L-Homocysteine | | 分子名称: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-14 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WON
 
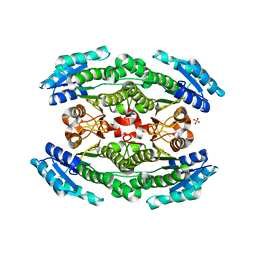 | | Crystal structure of acetoin dehydrogenase YohF from Salmonella typhimurium | | 分子名称: | CHLORIDE ION, SULFATE ION, YohF | | 著者 | Stogios, P.J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-25 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structure of acetoin dehydrogenase YohF from Salmonella typhimurium
To Be Published
|
|
7LGO
 
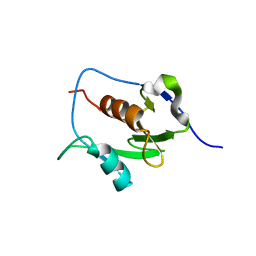 | | Crystal structure of the nucleic acid binding domain (NAB) of Nsp3 from SARS-CoV-2 | | 分子名称: | Non-structural protein 3 | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-01-20 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of the nucleic acid binding domain (NAB) of Nsp3 from SARS-CoV-2
To Be Published
|
|
5UE7
 
 | | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Phosphomannomutase | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-12-29 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state
To Be Published
|
|
5UF8
 
 | | Crystal structure of the ARF family small GTPase ARF2 from Candida albicans in complex with GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Potential ADP-ribosylation factor | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-01-03 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.872 Å) | | 主引用文献 | Crystal structure of the ARF family small GTPase ARF2 from Candida albicans in complex with GDP
To Be Published
|
|
