3NR1
 
 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | 分子名称: | HD domain-containing protein 3, MANGANESE (II) ION | | 著者 | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | 登録日 | 2010-06-30 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
3VCR
 
 | | Crystal structure of a putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase from Oleispira antarctica | | 分子名称: | PYRUVIC ACID, putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase | | 著者 | Stogios, P.J, Kagan, O, Di Leo, R, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-01-04 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
8FE9
 
 | | Crystal structure of Ack1 kinase K161Q mutant in complex with the selective inhibitor (R)-9b | | 分子名称: | Activated CDC42 kinase 1, N-[(1S)-1-benzyl-2-[(6-chloro-2-oxo-1H-quinolin-4-yl)methylamino]-2-oxo-ethyl]-4-hydroxy-2-oxo-1H-quinoline-6-carbo | | 著者 | Paung, Y, Kan, Y, Seeliger, M.S, Miller, W.T. | | 登録日 | 2022-12-06 | | 公開日 | 2023-03-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Biochemical Studies of Systemic Lupus Erythematosus-Associated Mutations in Nonreceptor Tyrosine Kinases Ack1 and Brk.
Biochemistry, 62, 2023
|
|
3V77
 
 | | Crystal structure of a putative fumarylacetoacetate isomerase/hydrolase from Oleispira antarctica | | 分子名称: | ACETATE ION, D(-)-TARTARIC ACID, Putative fumarylacetoacetate isomerase/hydrolase, ... | | 著者 | Stogios, P.J, Kagan, O, Di Leo, R, Bochkarev, A, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-12-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
6PI9
 
 | | Crystal structure of 16S rRNA methyltransferase RmtF in complex with S-Adenosyl-L-homocysteine | | 分子名称: | 16S rRNA (guanine(1405)-N(7))-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Stogios, P.J, Kim, Y, Evdokimova, E, Di Leo, R, Semper, C, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-06-26 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of 16S rRNA methylase RmtF in complex with S-Adenosyl-L-homocysteine
To be Published
|
|
2ZXX
 
 | | Crystal structure of Cdt1/geminin complex | | 分子名称: | DNA replication factor Cdt1, Geminin | | 著者 | Cho, Y, Lee, C, Hong, B.S, Choi, J.M. | | 登録日 | 2009-01-08 | | 公開日 | 2009-02-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for inhibition of the replication licensing factor Cdt1 by geminin
Nature, 430, 2004
|
|
8WMF
 
 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-1 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-10-03 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8WMD
 
 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-10-03 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8XLM
 
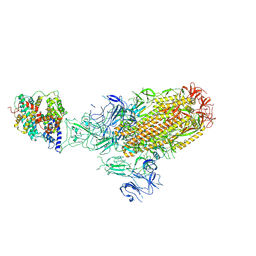 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-12-26 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8XLN
 
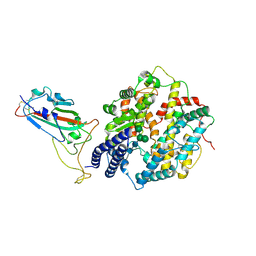 | | Structure of the SARS-CoV-2 EG.5.1 spike RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-12-26 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
7Z2K
 
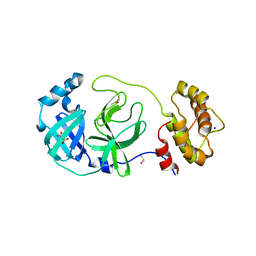 | | Crystal structure of SARS-CoV-2 Main Protease in orthorhombic space group p212121 | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | 登録日 | 2022-02-28 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
4UOY
 
 | |
4UOX
 
 | |
3ZGA
 
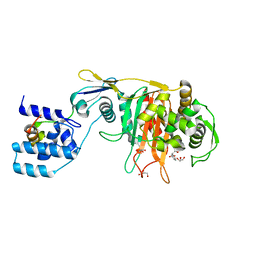 | | Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN 4 | | 著者 | Jeong, J.H, Kim, Y.G. | | 登録日 | 2012-12-17 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.005 Å) | | 主引用文献 | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZG7
 
 | |
6KEI
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 16-methoxy-11-methyl-6-[(pyridin-2-yl)methoxy]-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one | | 分子名称: | 16-methoxy-11-methyl-6-[(pyridin-2-yl)methoxy]-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | 著者 | Lee, B.I, Park, T.H. | | 登録日 | 2019-07-04 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6ISS
 
 | | Lignin peroxidase H8 triple mutant S49C/A67C/H239 | | 分子名称: | CALCIUM ION, Ligninase H8, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Seo, H, Son, H, Kim, K.-J. | | 登録日 | 2018-11-19 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Extra disulfide and ionic salt bridge improves the thermostability of lignin peroxidase H8 under acidic condition
Enzyme.Microb.Technol., 148, 2021
|
|
6XG3
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, at room temperature | | 分子名称: | CHLORIDE ION, Non-structural protein 3, PHOSPHATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
2HQR
 
 | |
2HQO
 
 | |
2HQN
 
 | |
7ROA
 
 | | Crystal structure of EntV136 from Enterococcus faecalis | | 分子名称: | EntV | | 著者 | Stogios, P.J, Evdokimova, E, Kim, Y, Garsin, D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2021-07-30 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-01-25 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural and functional analysis of EntV reveals a 12 amino acid fragment protective against fungal infections.
Nat Commun, 13, 2022
|
|
6MN5
 
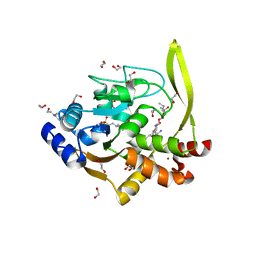 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, H154A mutant, in complex with gentamicin C1A | | 分子名称: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | 著者 | Stogios, P.J, Evdokimova, E, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-01 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
2Q7F
 
 | |
2PL2
 
 | | Crystal structure of TTC0263: a thermophilic TPR protein in Thermus thermophilus HB27 | | 分子名称: | Hypothetical conserved protein TTC0263 | | 著者 | Lim, H, Kim, K, Han, D, Oh, J. | | 登録日 | 2007-04-18 | | 公開日 | 2008-03-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of TTC0263, a thermophilic TPR protein from Thermus thermophilus HB27.
Mol.Cell, 24, 2007
|
|
