8Y9V
 
 | | ZIKV NS2B/NS3 protease | | 分子名称: | DAR-LYS-ORN-ARG, Serine protease NS3, Serine protease subunit NS2B | | 著者 | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H, Wang, Q.S. | | 登録日 | 2024-02-07 | | 公開日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
8YA1
 
 | |
8YA5
 
 | |
6NKK
 
 | | Structure of PhqE Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and premalbrancheamide | | 分子名称: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | 著者 | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | 登録日 | 2019-01-07 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
6NKI
 
 | | Structure of PhqB Reductase Domain from Penicillium fellutanum | | 分子名称: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NRPS | | 著者 | Dan, Q, Newmister, S.A, Smith, J.L, Sherman, D.H. | | 登録日 | 2019-01-07 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
6NKM
 
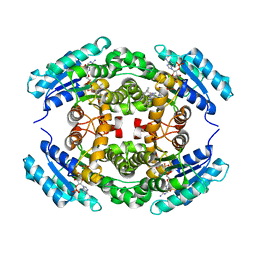 | | Structure of PhqE D166N Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and substrate | | 分子名称: | 3-{[2-(2-methylbut-3-en-2-yl)-1H-indol-3-yl]methyl}-8H-pyrrolo[1,2-a]pyrazin-5-ium-1-olate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | 著者 | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | 登録日 | 2019-01-07 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.896 Å) | | 主引用文献 | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
6NKH
 
 | |
8XAX
 
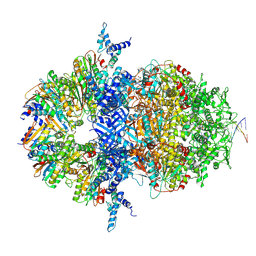 | |
8XAU
 
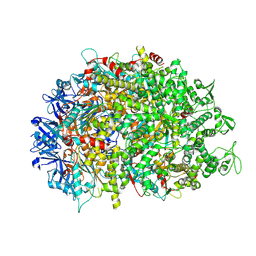 | | Cryo-EM structure of HerA | | 分子名称: | ATP-binding protein | | 著者 | Wang, Y, Deng, Z. | | 登録日 | 2023-12-05 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 2024
|
|
8XAV
 
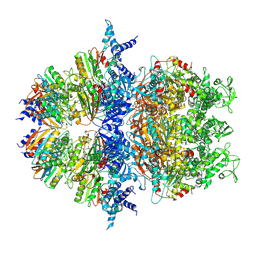 | |
8XAW
 
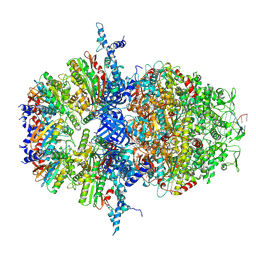 | |
8XAY
 
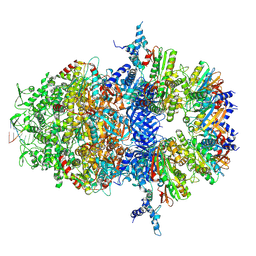 | |
4NWK
 
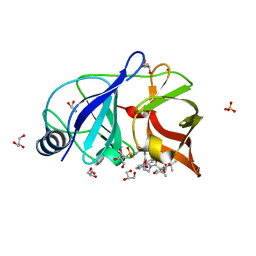 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-605339 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-n-((1r,2s)-1-((cyclopropylsulfonyl)carba moyl)-2-vinylcyclopropyl)-4-((6-methoxy-1-isoquinolinyl)ox y)-l-prolinamide | | 分子名称: | GLYCEROL, HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-4-[(6-methoxyisoquinolin-1-yl)oxy]-L-prolinamide, ... | | 著者 | Muckelbauer, J.K, Klei, H.E. | | 登録日 | 2013-12-06 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | 著者 | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | 登録日 | 2020-07-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.108 Å) | | 主引用文献 | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
4NWL
 
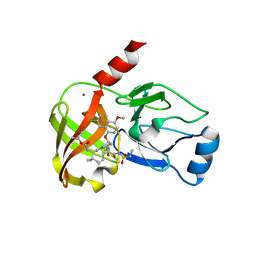 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-650032 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-4-((7-chloro-4-methoxy-1-isoquinolinyl)o xy)-n-((1r,2s)-1-((cyclopropylsulfonyl)carbamoyl)-2-vinylc yclopropyl)-l-prolinamide | | 分子名称: | HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, ZINC ION | | 著者 | Muckelbauer, J.K, Klei, H.E. | | 登録日 | 2013-12-06 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
3G4D
 
 | | Crystal Structure of (+)-delta-Cadinene Synthase from Gossypium arboreum and Evolutionary Divergence of Metal Binding Motifs for Catalysis | | 分子名称: | (+)-delta-cadinene synthase isozyme XC1, BETA-MERCAPTOETHANOL, GLYCEROL | | 著者 | Gennadios, H.A, Di Costanzo, L, Miller, D.J, Allemann, R.K, Christianson, D.W. | | 登録日 | 2009-02-03 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.403 Å) | | 主引用文献 | Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis.
Biochemistry, 48, 2009
|
|
6PVF
 
 | | Crystal structure of PhqK in complex with malbrancheamide B | | 分子名称: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Fraley, A.E, Smith, J.L, Sherman, D.H. | | 登録日 | 2019-07-20 | | 公開日 | 2020-01-22 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVH
 
 | | Crystal structure of PhqK in complex with paraherquamide K | | 分子名称: | (7aS,12S,12aR,13aS)-3,3,12,14,14-pentamethyl-3,7,11,12,13,13a,14,15-octahydro-8H,10H-7a,12a-(epiminomethano)indolizino[6,7-h]pyrano[3,2-a]carbazol-16-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Fraley, A.E, Smith, J.L, Sherman, D.H. | | 登録日 | 2019-07-20 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVG
 
 | |
6PVI
 
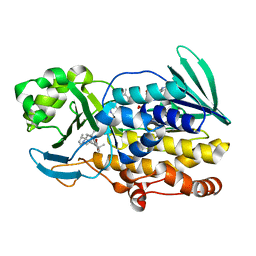 | | Crystal structure of PhqK in complex with paraherquamide L | | 分子名称: | (8aS,13S,13aR,14aS)-4,4,13,15,15-pentamethyl-12,13,14,14a,15,16-hexahydro-4H,8H,9H,11H-8a,13a-(epiminomethano)[1,4]dioxepino[2,3-a]indolizino[6,7-h]carbazol-17-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Fraley, A.E, Smith, J.L, Sherman, D.H. | | 登録日 | 2019-07-20 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.093 Å) | | 主引用文献 | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVJ
 
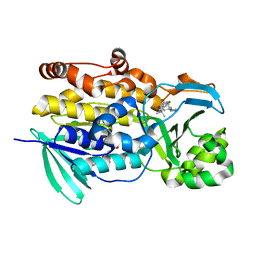 | | Crystal structure of PhqK in complex with malbrancheamide C | | 分子名称: | (5aS,12aS,13aS)-9-bromo-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Fraley, A.E, Smith, J.L, Sherman, D.H. | | 登録日 | 2019-07-20 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
3G4F
 
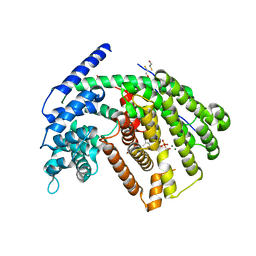 | | Crystal Structure of (+)- -Cadinene Synthase from Gossypium arboreum in complex with 2-fluorofarnesyl diphosphate | | 分子名称: | (+)-delta-cadinene synthase isozyme XC1, (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, BETA-MERCAPTOETHANOL, ... | | 著者 | Gennadios, H.A, Di Costanzo, L, Christianson, D.W. | | 登録日 | 2009-02-03 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.651 Å) | | 主引用文献 | Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis.
Biochemistry, 48, 2009
|
|
8GWO
 
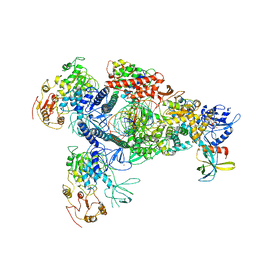 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWG
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWI
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
