1J2U
 
 | | Creatininase Zn | | 分子名称: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | 著者 | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | 登録日 | 2003-01-11 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1IDM
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | 登録日 | 1995-05-19 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1WNI
 
 | | Crystal Structure of H2-Proteinase | | 分子名称: | Trimerelysin II, ZINC ION | | 著者 | Kumasaka, T, Yamamoto, M, Moriyama, H, Tanaka, N, Sato, M, Katsube, Y, Yamakawa, Y, Omori-Satoh, T, Iwanaga, S, Ueki, T. | | 登録日 | 2004-08-04 | | 公開日 | 2004-08-17 | | 最終更新日 | 2019-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of H2-proteinase from the venom of Trimeresurus flavoviridis.
J.Biochem., 119, 1996
|
|
1XER
 
 | | STRUCTURE OF FERREDOXIN | | 分子名称: | FE3-S4 CLUSTER, FERREDOXIN, ZINC ION | | 著者 | Fujii, T, Hata, Y, Moriyama, H, Wakagi, T, Tanaka, N, Oshima, T. | | 登録日 | 1996-08-28 | | 公開日 | 1997-09-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Novel zinc-binding centre in thermoacidophilic archaeal ferredoxins.
Nat.Struct.Biol., 3, 1996
|
|
8JNW
 
 | | PfDXR - Mn2+ - NADPH - MAMK89 quaternary complex | | 分子名称: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | 著者 | Takada, S, Sakamoto, Y, Tanaka, N. | | 登録日 | 2023-06-08 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Reverse N -Substituted Hydroxamic Acid Derivatives of Fosmidomycin Target a Previously Unknown Subpocket of 1-Deoxy-d-xylulose 5-Phosphate Reductoisomerase (DXR).
Acs Infect Dis., 10, 2024
|
|
8JNV
 
 | | PfDXR - Mn2+ - MAMK89 ternary complex | | 分子名称: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | 著者 | Takada, S, Sakamoto, Y, Tanaka, N. | | 登録日 | 2023-06-06 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Reverse N -Substituted Hydroxamic Acid Derivatives of Fosmidomycin Target a Previously Unknown Subpocket of 1-Deoxy-d-xylulose 5-Phosphate Reductoisomerase (DXR).
Acs Infect Dis., 10, 2024
|
|
1XAC
 
 | |
1XAD
 
 | |
1WQS
 
 | | Crystal structure of Norovirus 3C-like protease | | 分子名称: | 3C-like protease, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | 著者 | Nakamura, K, Someya, Y, Kumasaka, T, Tanaka, N. | | 登録日 | 2004-10-01 | | 公開日 | 2005-10-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A norovirus protease structure provides insights into active and substrate binding site integrity
J.Virol., 79, 2005
|
|
5YP4
 
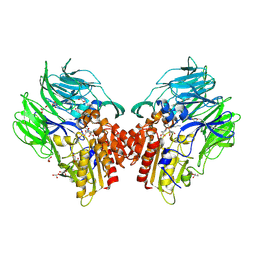 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 | | 分子名称: | Dipeptidyl aminopeptidase 4, GLYCEROL, LYSINE, ... | | 著者 | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5XXM
 
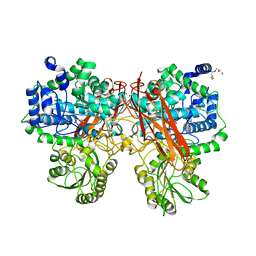 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone | | 分子名称: | D-glucono-1,5-lactone, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | 著者 | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | 登録日 | 2017-07-04 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5YP1
 
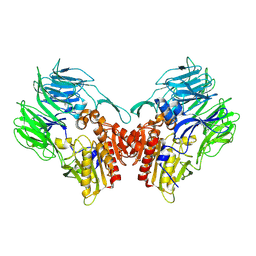 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 | | 分子名称: | Dipeptidyl aminopeptidase 4, GLYCEROL | | 著者 | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP2
 
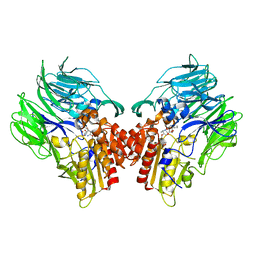 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 | | 分子名称: | (2S,5R)-1-[2-[[1-(hydroxymethyl)cyclopentyl]amino]ethanoyl]pyrrolidine-2,5-dicarbonitrile, Dipeptidyl aminopeptidase 4, GLYCEROL | | 著者 | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP3
 
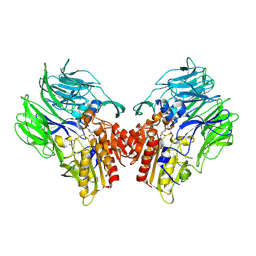 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana | | 分子名称: | Dipeptidyl aminopeptidase 4, GLYCEROL, ISOLEUCINE, ... | | 著者 | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | 分子名称: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | 著者 | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | 登録日 | 2017-12-18 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
5XXL
 
 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | 著者 | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | 登録日 | 2017-07-04 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5XXO
 
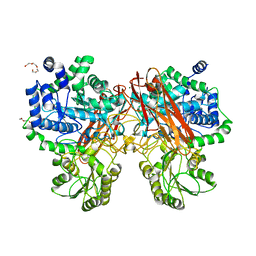 | | Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | 著者 | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | 登録日 | 2017-07-04 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5XXN
 
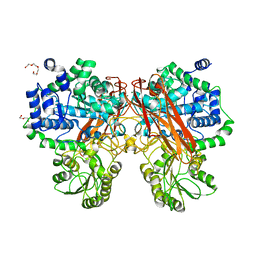 | | Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | 著者 | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahashi, Y, Sugimono, N, Nakai, H, Taguchi, H. | | 登録日 | 2017-07-04 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
1VEC
 
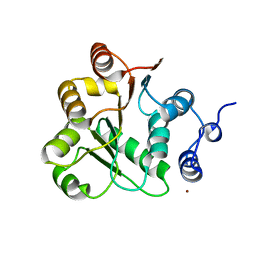 | | Crystal structure of the N-terminal domain of rck/p54, a human DEAD-box protein | | 分子名称: | ATP-dependent RNA helicase p54, L(+)-TARTARIC ACID, ZINC ION | | 著者 | Hogetsu, K, Matsui, T, Yukihiro, Y, Tanaka, M, Sato, T, Kumasaka, T, Tanaka, N. | | 登録日 | 2004-03-29 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural insight of human DEAD-box protein rck/p54 into its substrate recognition with conformational changes
Genes Cells, 11, 2006
|
|
1XAB
 
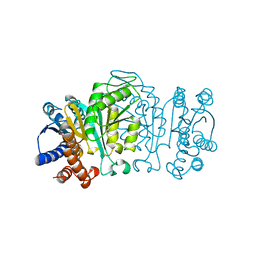 | |
1XAA
 
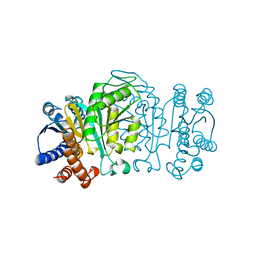 | |
1VBU
 
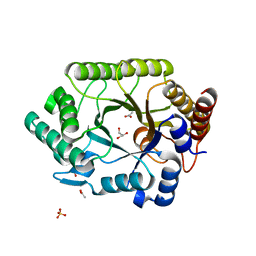 | | Crystal structure of native xylanase 10B from Thermotoga maritima | | 分子名称: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | 著者 | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | 登録日 | 2004-03-02 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
1VBR
 
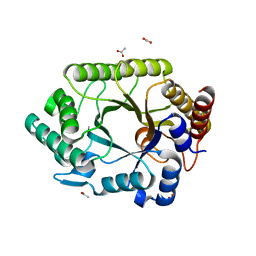 | | Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose | | 分子名称: | ACETIC ACID, alpha-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase B | | 著者 | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | 登録日 | 2004-03-02 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
1UJ0
 
 | | Crystal Structure of STAM2 SH3 domain in complex with a UBPY-derived peptide | | 分子名称: | PHOSPHATE ION, deubiquitinating enzyme UBPY, signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | | 著者 | Kaneko, T, Kumasaka, T, Ganbe, T, Sato, T, Miyazawa, K, Kitamura, N, Tanaka, N. | | 登録日 | 2003-07-24 | | 公開日 | 2003-12-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain.
J.Biol.Chem., 278, 2003
|
|
1V7Z
 
 | | creatininase-product complex | | 分子名称: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | 著者 | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | 登録日 | 2003-12-26 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
