4G2T
 
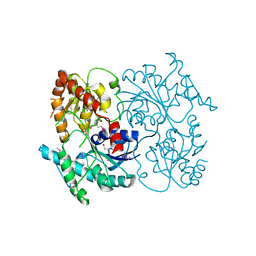 | | Crystal Structure of Streptomyces sp. SF2575 glycosyltransferase SsfS6, complexed with thymidine diphosphate | | 分子名称: | SsfS6, THYMIDINE-5'-DIPHOSPHATE | | 著者 | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-07-12 | | 公開日 | 2012-07-25 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.405 Å) | | 主引用文献 | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
4HPV
 
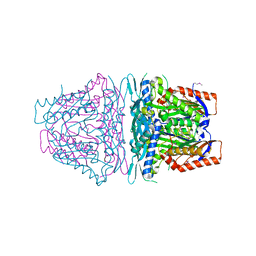 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus | | 分子名称: | S-adenosylmethionine synthase | | 著者 | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-10-24 | | 公開日 | 2012-11-14 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.214 Å) | | 主引用文献 | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
5EEH
 
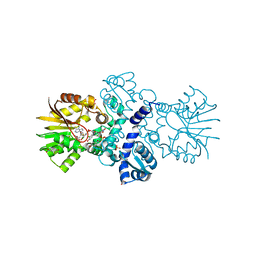 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 2-chloro-4-nitrophenol | | 分子名称: | 2-chloranyl-4-nitro-phenol, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-10-22 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
5EEG
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with tetrazole-SAH | | 分子名称: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[[(3~{S})-3-azanyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)propyl]sulfanylmethyl]oxolane-3,4-diol, Carminomycin 4-O-methyltransferase DnrK | | 著者 | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-10-22 | | 公開日 | 2015-12-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.255 Å) | | 主引用文献 | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
2KMW
 
 | |
4P8E
 
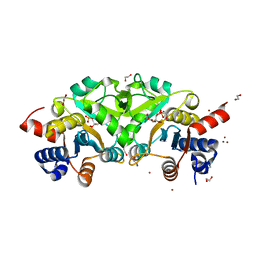 | | Structure of ribB complexed with substrate (Ru5P) and metal ions | | 分子名称: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE, ... | | 著者 | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | 登録日 | 2014-03-31 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6C
 
 | | Structure of ribB complexed with inhibitor 4PEH | | 分子名称: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID | | 著者 | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | 登録日 | 2014-03-24 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
5HOQ
 
 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | 分子名称: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | 著者 | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | 登録日 | 2016-01-19 | | 公開日 | 2016-03-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.793 Å) | | 主引用文献 | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
4P77
 
 | | Structure of ribB complexed with substrate Ru5P | | 分子名称: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, RIBULOSE-5-PHOSPHATE | | 著者 | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | 登録日 | 2014-03-26 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6D
 
 | | Structure of ribB complexed with PO4 ion | | 分子名称: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, PHOSPHATE ION | | 著者 | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | 登録日 | 2014-03-24 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6P
 
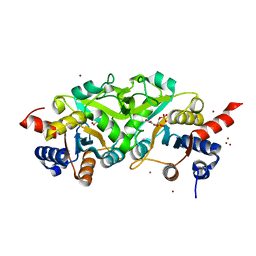 | | Structure of ribB complexed with inhibitor (4PEH) and metal ions | | 分子名称: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, ZINC ION | | 著者 | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | 登録日 | 2014-03-25 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.862 Å) | | 主引用文献 | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P8J
 
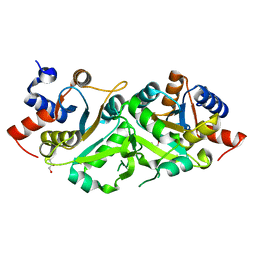 | | Structure of ribB | | 分子名称: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL | | 著者 | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | 登録日 | 2014-03-31 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
2LUZ
 
 | | Solution NMR Structure of CalU16 from Micromonospora echinospora, Northeast Structural Genomics Consortium (NESG) Target MiR12 | | 分子名称: | CalU16 | | 著者 | Ramelot, T.A, Yang, Y, Lee, H, Pederson, K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Wrobel, R.L, Bingman, C.A, Singh, S, Thorson, J.S, Prestegard, J.H, Montelione, G.T, Phillips Jr, G.N, Kennedy, M.A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-06-22 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | 分子名称: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | 著者 | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-08-12 | | 公開日 | 2013-09-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
1S6W
 
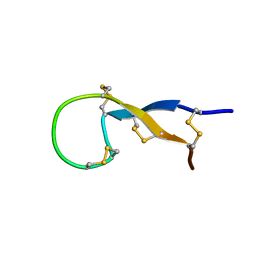 | | Solution Structure of hybrid white striped bass hepcidin | | 分子名称: | Hepcidin | | 著者 | Babon, J.J, Singh, S, Pennington, M.W, Norton, R.S, Westerman, M.E. | | 登録日 | 2004-01-28 | | 公開日 | 2004-12-14 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections.
J.Biol.Chem., 280, 2005
|
|
1L4U
 
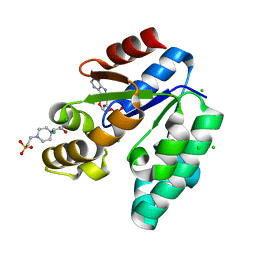 | | CRYSTAL STRUCTURE OF SHIKIMATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MGADP AND PT(II) AT 1.8 ANGSTROM RESOLUTION | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Gu, Y, Reshetnikova, L, Li, Y, Wu, Y, Yan, H, Singh, S, Ji, X. | | 登録日 | 2002-03-05 | | 公開日 | 2002-06-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of shikimate kinase from Mycobacterium tuberculosis reveals the dynamic role of the LID domain in catalysis.
J.Mol.Biol., 319, 2002
|
|
1LB4
 
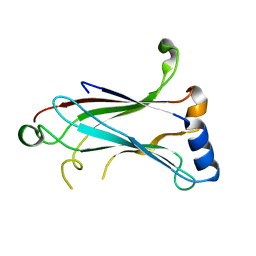 | | TRAF6 apo structure | | 分子名称: | TNF receptor-associated factor 6 | | 著者 | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | 登録日 | 2002-04-02 | | 公開日 | 2002-07-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1LB5
 
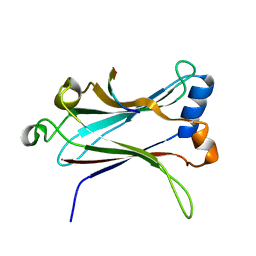 | | TRAF6-RANK Complex | | 分子名称: | TNF receptor-associated factor 6, receptor activator of nuclear factor-kappa B | | 著者 | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | 登録日 | 2002-04-02 | | 公開日 | 2002-07-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1L4Y
 
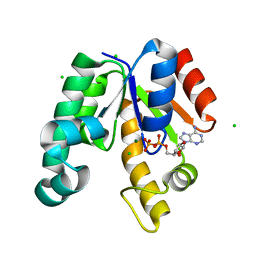 | | CRYSTAL STRUCTURE OF SHIKIMATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MGADP AT 2.0 ANGSTROM RESOLUTION | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Gu, Y, Reshetnikova, L, Li, Y, Wu, Y, Yan, H, Singh, S, Ji, X. | | 登録日 | 2002-03-06 | | 公開日 | 2002-06-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of shikimate kinase from Mycobacterium tuberculosis reveals the dynamic role of the LID domain in catalysis.
J.Mol.Biol., 319, 2002
|
|
1LB6
 
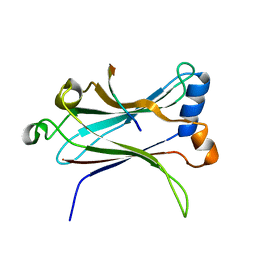 | | TRAF6-CD40 Complex | | 分子名称: | CD40 antigen, TNF receptor-associated factor 6 | | 著者 | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | 登録日 | 2002-04-02 | | 公開日 | 2002-07-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1M7Q
 
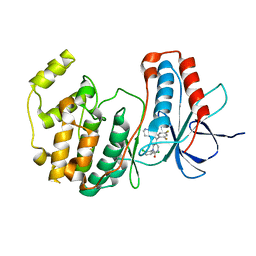 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | 分子名称: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | 著者 | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | 登録日 | 2002-07-22 | | 公開日 | 2002-12-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
3LST
 
 | | Crystal Structure of CalO1, Methyltransferase in Calicheamicin Biosynthesis, SAH bound form | | 分子名称: | 1,2-ETHANEDIOL, CalO1 Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2010-02-12 | | 公開日 | 2010-03-02 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural characterization of CalO1: a putative orsellinic acid methyltransferase in the calicheamicin-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OTI
 
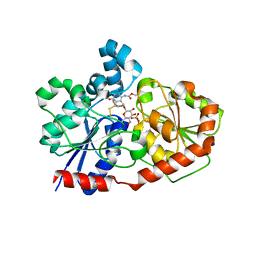 | | Crystal Structure of CalG3, Calicheamicin Glycostyltransferase, TDP and calicheamicin T0 bound form | | 分子名称: | CHLORIDE ION, CalG3, Calicheamicin T0, ... | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2010-09-11 | | 公開日 | 2010-12-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.597 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTH
 
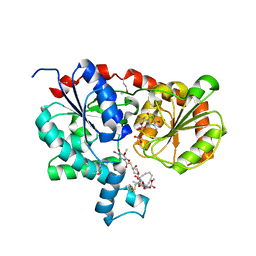 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP and calicheamicin alpha3I bound form | | 分子名称: | CalG1, Calicheamicin alpha3I, THYMIDINE-5'-DIPHOSPHATE | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2010-09-11 | | 公開日 | 2010-12-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTG
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form | | 分子名称: | CHLORIDE ION, CalG1, THYMIDINE-5'-DIPHOSPHATE | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2010-09-11 | | 公開日 | 2010-12-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
