4N90
 
 | |
4ES4
 
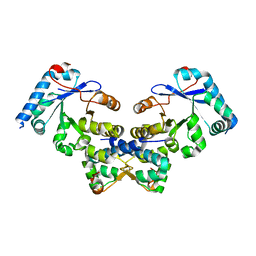 | | Crystal structure of YdiV and FlhD complex | | 分子名称: | Flagellar transcriptional regulator FlhD, Putative cyclic di-GMP regulator CdgR | | 著者 | Li, B, Gu, L. | | 登録日 | 2012-04-22 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
4I10
 
 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates | | 分子名称: | 2-{(1S)-1-[(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)amino]-2-phenylethyl}pyrido[4,3-d]pyrimidin-4(1H)-one, Beta-secretase 1, ZINC ION | | 著者 | Lougheed, J.C, Brecht, E, Yao, N.H. | | 登録日 | 2012-11-19 | | 公開日 | 2013-03-06 | | 最終更新日 | 2013-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I0D
 
 | |
4I12
 
 | | Design and synthesis of thiophene dihydroisoquinolins as novel BACE-1 inhibitors | | 分子名称: | 2-{(1S)-1-{[(1Z)-6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1(2H)-ylidene]amino}-2-[2-propyl-4-(1H-pyrazol-4-yl)thiophen-3-yl]ethyl}pyrimidin-4(5H)-one, Beta-secretase 1, SODIUM ION, ... | | 著者 | Lougheed, J.C, Brecht, E, Yao, N.H. | | 登録日 | 2012-11-19 | | 公開日 | 2013-03-06 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Design and synthesis of thiophene dihydroisoquinolines as novel BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I0G
 
 | |
4HZT
 
 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates | | 分子名称: | 3-{(1S)-1-[(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)amino]-2-phenylethyl}-1,2,4-oxadiazol-5(2H)-one, Beta-secretase 1, ZINC ION | | 著者 | Yao, N, Brecht, E. | | 登録日 | 2012-11-15 | | 公開日 | 2013-03-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I1C
 
 | | Design and synthesis of thiophene dihydroisoquinolins as novel BACE-1 inhibitors | | 分子名称: | BETA-SECRETASE 1, N-(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)-3-[2-propyl-4-(1H-pyrazol-4-yl)thiophen-3-yl]-L-alanine, ZINC ION | | 著者 | Lougheed, J.C, Brecht, E, Yao, N.H. | | 登録日 | 2012-11-20 | | 公開日 | 2013-03-06 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design and synthesis of thiophene dihydroisoquinolines as novel BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
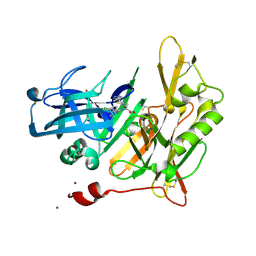
4I0E
 
 | |
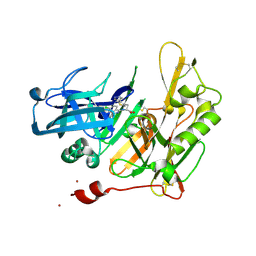
4I0F
 
 | |
9DU7
 
 | |
8T48
 
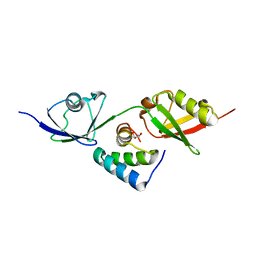 | |
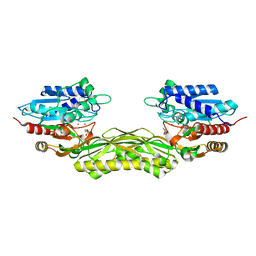
8C46
 
 | |
7K3C
 
 | | SGMGGIT segment 58-64 from Keratin-8 | | 分子名称: | ETHANOL, SGMGGIT segment 58-64 from the low complexity domain of Keratin-8, TRIETHYLENE GLYCOL | | 著者 | Murray, K.A, Sawaya, M.R, Eisenberg, D.S. | | 登録日 | 2020-09-11 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Identifying amyloid-related diseases by mapping mutations in low-complexity protein domains to pathologies.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7K3Y
 
 | |
7K3X
 
 | |
5V45
 
 | |
5V46
 
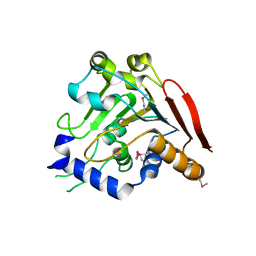 | | Crystal structure of the I113M, F270M, K291M, L308M mutant of SR1 domain of human sacsin | | 分子名称: | Sacsin | | 著者 | Menade, M, Kozlov, G, Gehring, K. | | 登録日 | 2017-03-08 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
5V44
 
 | |
7S5U
 
 | |
5VRG
 
 | |
5EK0
 
 | | Human Nav1.7-VSD4-NavAb in complex with GX-936. | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-cyano-4-[2-[2-(1-ethylazetidin-3-yl)pyrazol-3-yl]-4-(trifluoromethyl)phenoxy]-~{N}-(1,2,4-thiadiazol-5-yl)benzenesulfonamide, Chimera of bacterial Ion transport protein and human Sodium channel protein type 9 subunit alpha | | 著者 | Ahuja, S, Mukund, S, Starovasnik, M.A, Koth, C.M, Payandeh, J. | | 登録日 | 2015-11-03 | | 公開日 | 2015-12-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.53 Å) | | 主引用文献 | Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist.
Science, 350, 2015
|
|
5VRH
 
 | | Apolipoprotein N-acyltransferase C387S active site mutant | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, ... | | 著者 | Murray, J.M, Noland, C.L. | | 登録日 | 2017-05-10 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.137 Å) | | 主引用文献 | Structural insights into lipoprotein N-acylation by Escherichia coli apolipoprotein N-acyltransferase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1WDF
 
 | | crystal structure of MHV spike protein fusion core | | 分子名称: | E2 glycoprotein | | 著者 | Xu, Y, Liu, Y, Lou, Z, Qin, L, Li, X, Bai, Z, Tien, P, Gao, G.F, Rao, Z. | | 登録日 | 2004-05-14 | | 公開日 | 2004-06-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for Coronavirus-mediated Membrane Fusion: CRYSTAL STRUCTURE OF MOUSE HEPATITIS VIRUS SPIKE PROTEIN FUSION CORE
J.Biol.Chem., 279, 2004
|
|
1WDG
 
 | | crystal structure of MHV spike protein fusion core | | 分子名称: | E2 glycoprotein | | 著者 | Xu, Y, Liu, Y, Lou, Z, Qin, L, Li, X, Bai, Z, Tien, P, Gao, G.F, Rao, Z. | | 登録日 | 2004-05-14 | | 公開日 | 2004-06-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Structural Basis for Coronavirus-mediated Membrane Fusion: CRYSTAL STRUCTURE OF MOUSE HEPATITIS VIRUS SPIKE PROTEIN FUSION CORE
J.Biol.Chem., 279, 2004
|
|
