5U88
 
 | | Crystal structure of a MerB-triimethyllead complex. | | 分子名称: | ACETATE ION, Alkylmercury lyase, Trimethyllead bromide | | 著者 | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | 登録日 | 2016-12-14 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U7B
 
 | | Crystal structure of a the tin-bound form of MerB formed from Diethyltin. | | 分子名称: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | 著者 | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | 登録日 | 2016-12-12 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U7C
 
 | | Crystal structure of the lead-bound form of MerB formed from diethyllead. | | 分子名称: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | 著者 | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | 登録日 | 2016-12-12 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U82
 
 | | Crystal structure of a MerB-triethyltin complex | | 分子名称: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | 著者 | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | 登録日 | 2016-12-13 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U4K
 
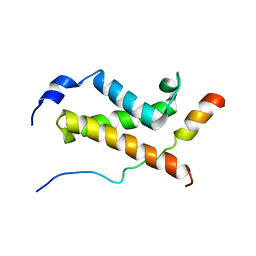 | | NMR structure of the complex between the KIX domain of CBP and the transactivation domain 1 of p65 | | 分子名称: | CREB-binding protein, Transcription factor p65 | | 著者 | Lecoq, L, Raiola, L, Chabot, P.R, Cyr, N, Arseneault, G, Omichinski, J.G. | | 登録日 | 2016-12-05 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
5U79
 
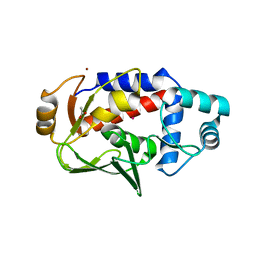 | | Crystal structure of a complex formed between MerB and Dimethyltin | | 分子名称: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | 著者 | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | 登録日 | 2016-12-12 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.604 Å) | | 主引用文献 | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U83
 
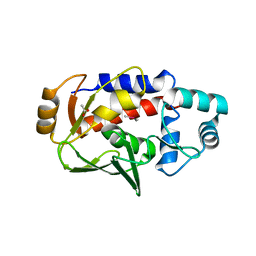 | | Crystal structure of a MerB-trimethytin complex. | | 分子名称: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | 著者 | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | 登録日 | 2016-12-13 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.609 Å) | | 主引用文献 | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
3SAK
 
 | |
2LOX
 
 | |
2MKR
 
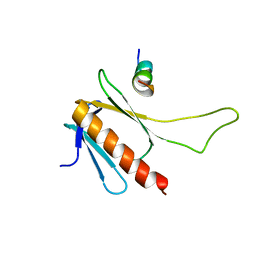 | | Structural Characterization of a Complex Between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 subunit of TFIIH. | | 分子名称: | Epstein-Barr nuclear antigen 2, RNA polymerase II transcription factor B subunit 1 | | 著者 | Chabot, P.R, Raiola, L, Lussier-Price, M, Morse, T, Arseneault, G, Archambault, J, Omichinski, J. | | 登録日 | 2014-02-12 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH.
Plos Pathog., 10, 2014
|
|
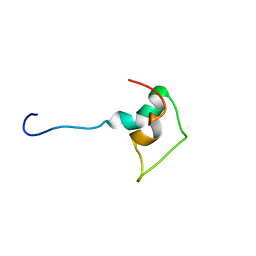
1BAL
 
 | |
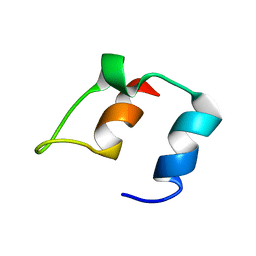
1BBL
 
 | |
8T2N
 
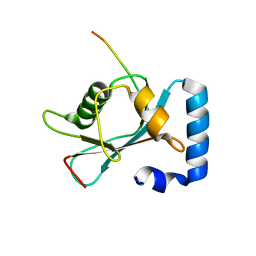 | | Crystal structure of GABARAP in complex with the LIR of NSs3 | | 分子名称: | Gamma-aminobutyric acid receptor-associated protein, Non-structural protein S | | 著者 | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | 登録日 | 2023-06-06 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8T2M
 
 | | Crystal structure of GABARAP in complex with the LIR of NSs4 | | 分子名称: | Non-structural protein S,Gamma-aminobutyric acid receptor-associated protein chimera | | 著者 | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | 登録日 | 2023-06-06 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8T2L
 
 | | Crystal structure of LC3A in complex with the LIR of NSs4 | | 分子名称: | Non-structural protein S,Microtubule-associated proteins 1A/1B light chain 3A chimera | | 著者 | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | 登録日 | 2023-06-06 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
