2DOQ
 
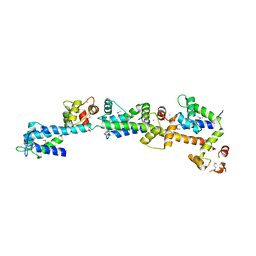 | | crystal structure of Sfi1p/Cdc31p complex | | 分子名称: | CALCIUM ION, Cell division control protein 31, SFI1p | | 著者 | Li, S, Sandercock, A.M, Conduit, P.T, Robinson, C.V, Williams, R.L, Kilmartin, J.V. | | 登録日 | 2006-05-03 | | 公開日 | 2006-06-27 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J.Cell Biol., 173, 2006
|
|
8K3Y
 
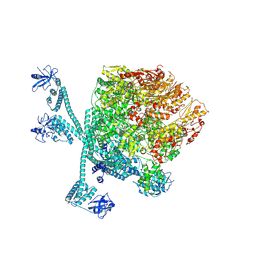 | | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | 著者 | Li, S, Hsieh, K.Y, Kuo, C.I, Zhang, K, Chang, C.I. | | 登録日 | 2023-07-17 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (4.42 Å) | | 主引用文献 | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
6IN9
 
 | |
6IN7
 
 | |
6IN8
 
 | | Crystal structure of MucB | | 分子名称: | Sigma factor AlgU regulatory protein MucB | | 著者 | Li, S, Zhang, Q, Bartlam, M. | | 登録日 | 2018-10-24 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
7F1E
 
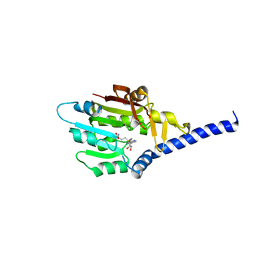 | | Structure of METTL6 bound with SAM | | 分子名称: | S-ADENOSYLMETHIONINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | 著者 | Li, S, Liao, S, Xu, C. | | 登録日 | 2021-06-09 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.589 Å) | | 主引用文献 | Structural basis for METTL6-mediated m3C RNA methylation.
Biochem.Biophys.Res.Commun., 589, 2021
|
|
7FD4
 
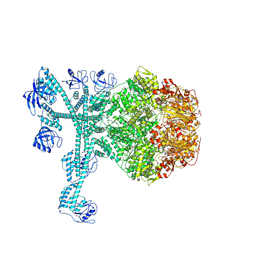 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | 登録日 | 2021-07-16 | | 公開日 | 2021-11-03 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7FD5
 
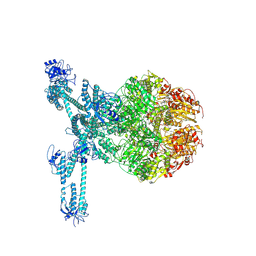 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | 登録日 | 2021-07-16 | | 公開日 | 2021-11-03 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7FIZ
 
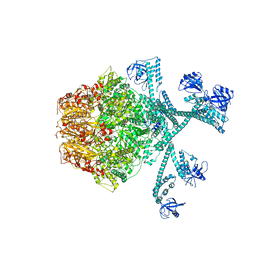 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 3) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-08-01 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIE
 
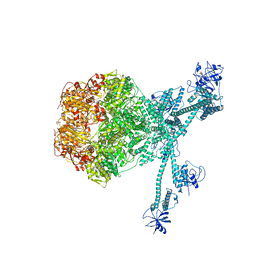 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-07-31 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FID
 
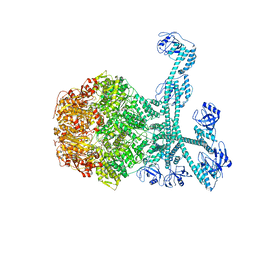 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon proteasecomplex (conformation 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-07-31 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7YGA
 
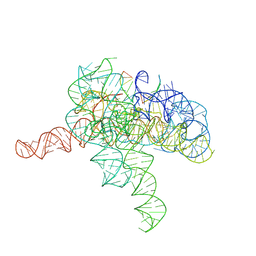 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.35 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGC
 
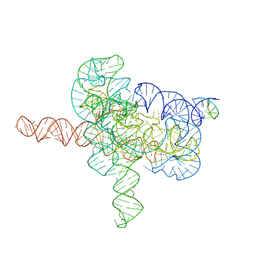 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*UP*UP*AP*AP*CP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-04-05 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YG8
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 5 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (387-MER), RNA (5'-R(*CP*CP*CP*UP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGD
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (384-MER), RNA (5'-R(*CP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGB
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*UP*UP*AP*AP*CP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YG9
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (391-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | 分子名称: | RNA (388-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | 分子名称: | RNA (388-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | 分子名称: | RNA (387-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | 分子名称: | RNA (388-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (4.01 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8JON
 
 | | Structure of a synthetic circadian clock protein KaiC mutant of cyanobacteria Synechococcus elongatus PCC 7942 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, ... | | 著者 | Jia, X, Zhang, Q, Li, S, Guo, J. | | 登録日 | 2023-06-07 | | 公開日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Adaptation of ancient cyanobacterial clock to the day length ~ 0.95 Ga ago
To Be Published
|
|
3U28
 
 | |
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | 著者 | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | 登録日 | 2020-04-28 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
8HVS
 
 | |
