3KR9
 
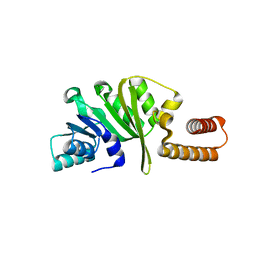 | |
3KU1
 
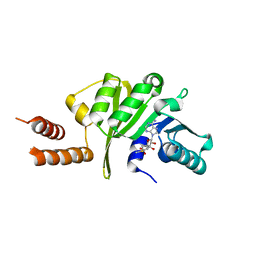 | |
1PCV
 
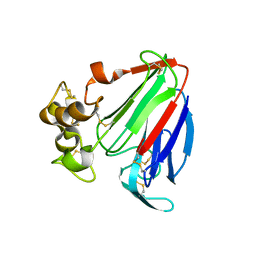 | | Crystal structure of osmotin, a plant antifungal protein | | 分子名称: | osmotin | | 著者 | Min, K, Ha, S.C, Yun, D.-J, Bressan, R.A, Kim, K.K. | | 登録日 | 2003-05-16 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of osmotin, a plant antifungal protein
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3U7L
 
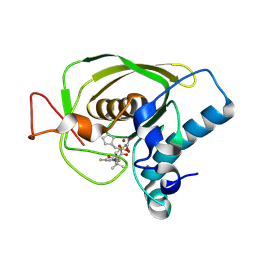 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | 分子名称: | (S)-N-(cyclopentylmethyl)-2-(3-(3,5-difluorophenyl)ureido)-N-(2-(hydroxyamino)-2-oxoethyl)-3,3-dimethylbutanamide, Peptide deformylase, ZINC ION | | 著者 | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | 登録日 | 2011-10-14 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7M
 
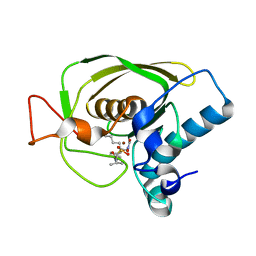 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | 分子名称: | N-((2R,4S)-2-butyl-4-(3-(2-fluorophenyl)ureido)-5-methyl-3-oxohexyl)-N-hydroxyformamide, Peptide deformylase, ZINC ION | | 著者 | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | 登録日 | 2011-10-14 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7K
 
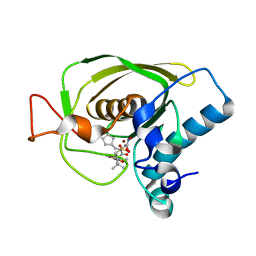 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | 分子名称: | (S)-N-(cyclopentylmethyl)-N-(2-(hydroxyamino)-2-oxoethyl)-2-(3-(2-methoxyphenyl)ureido)-3,3-dimethylbutanamide, Peptide deformylase, ZINC ION | | 著者 | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | 登録日 | 2011-10-14 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7N
 
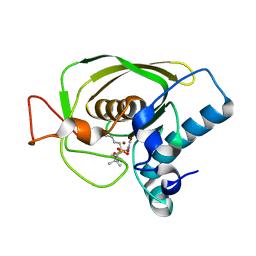 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | 分子名称: | N-((2R,4S)-2-butyl-5-methyl-4-(3-(5-methylpyridin-2-yl)ureido)-3-oxohexyl)-N-hydroxyformamide, Peptide deformylase, ZINC ION | | 著者 | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | 登録日 | 2011-10-14 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4TX1
 
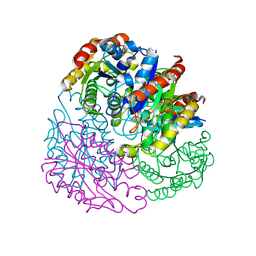 | |
6IYM
 
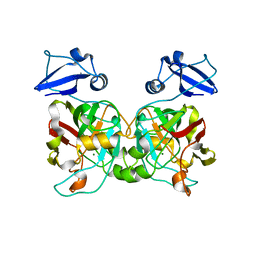 | |
5LS0
 
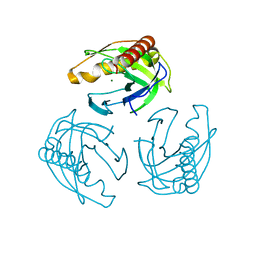 | | Crystal structure of Inorganic Pyrophosphatase PPA1 from Arabidopsis thaliana | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Soluble inorganic pyrophosphatase 1 | | 著者 | Grzechowiak, M, Sikorski, M, Jaskolski, M. | | 登録日 | 2016-08-22 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Crystal structures of plant inorganic pyrophosphatase, an enzyme with a moonlighting autoproteolytic activity.
Biochem.J., 476, 2019
|
|
1JTB
 
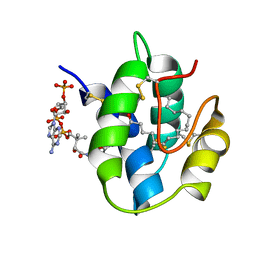 | | LIPID TRANSFER PROTEIN COMPLEXED WITH PALMITOYL COENZYME A, NMR, 16 STRUCTURES | | 分子名称: | COENZYME A, LIPID TRANSFER PROTEIN, PALMITIC ACID | | 著者 | Lerche, M.H, Kragelund, B.B, Bech, L.M, Poulsen, F.M. | | 登録日 | 1996-12-03 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-06-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Barley lipid-transfer protein complexed with palmitoyl CoA: the structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands.
Structure, 5, 1997
|
|
1W00
 
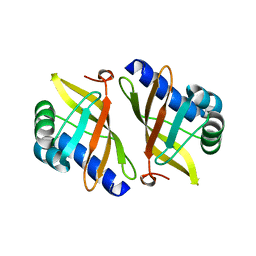 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 2004-05-30 | | 公開日 | 2005-05-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
1W01
 
 | |
1W02
 
 | |
1MZM
 
 | |
6JZL
 
 | |
8HEA
 
 | | Esterase2 (EaEst2) from Exiguobacterium antarcticum | | 分子名称: | Thermostable carboxylesterase Est30 | | 著者 | Hwang, J, Lee, J.H. | | 登録日 | 2022-11-08 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural and Biochemical Insights into Bis(2-hydroxyethyl) Terephthalate Degrading Carboxylesterase Isolated from Psychrotrophic Bacterium Exiguobacterium antarcticum.
Int J Mol Sci, 24, 2023
|
|
2G97
 
 | |
2G96
 
 | |
2G95
 
 | |
1VJS
 
 | |
1DMQ
 
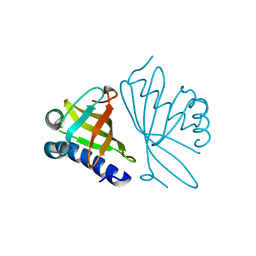 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 1999-12-14 | | 公開日 | 2000-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1OPY
 
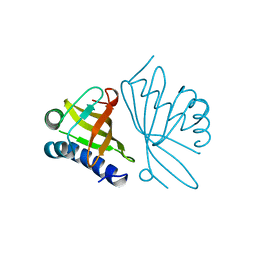 | | KSI | | 分子名称: | DELTA5-3-KETOSTEROID IOSMERASE | | 著者 | Kim, S.-W, Cha, S.-S, Cho, H.-S, Kim, J.-S, Ha, N.-C, Cho, M.-J, Choi, K.-Y, Oh, B.-H. | | 登録日 | 1997-05-23 | | 公開日 | 1998-04-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-resolution crystal structures of delta5-3-ketosteroid isomerase with and without a reaction intermediate analogue.
Biochemistry, 36, 1997
|
|
1DMM
 
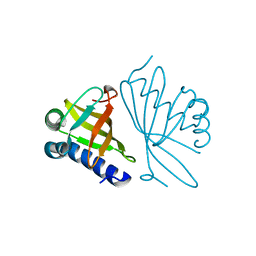 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 1999-12-14 | | 公開日 | 2000-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMN
 
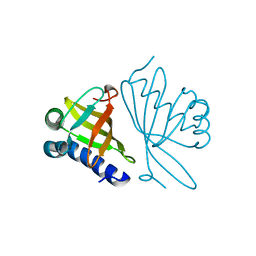 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F/Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 1999-12-14 | | 公開日 | 2000-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
