2H4X
 
 | |
2H5E
 
 | |
2HHJ
 
 | | Human bisphosphoglycerate mutase complexed with 2,3-bisphosphoglycerate (15 days) | | 分子名称: | (2R)-2,3-diphosphoglyceric acid, 3-PHOSPHOGLYCERIC ACID, Bisphosphoglycerate mutase, ... | | 著者 | Wang, Y, Gong, W. | | 登録日 | 2006-06-28 | | 公開日 | 2006-10-24 | | 最終更新日 | 2016-07-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase.
J.Biol.Chem., 281, 2006
|
|
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | 分子名称: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | 著者 | Zhao, S, Huang, J, Li, H. | | 登録日 | 2017-01-07 | | 公開日 | 2017-08-16 | | 最終更新日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.297 Å) | | 主引用文献 | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | 分子名称: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | 著者 | Zhao, S, Li, H. | | 登録日 | 2017-01-07 | | 公開日 | 2017-08-16 | | 最終更新日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.703 Å) | | 主引用文献 | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3D45
 
 | | Crystal structure of mouse PARN in complex with m7GpppG | | 分子名称: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Poly(A)-specific ribonuclease PARN | | 著者 | Wu, M, Song, H. | | 登録日 | 2008-05-13 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis of m(7)GpppG binding to poly(A)-specific ribonuclease.
Structure, 17, 2009
|
|
3EIJ
 
 | | Crystal structure of Pdcd4 | | 分子名称: | Programmed cell death protein 4 | | 著者 | Loh, P.G. | | 登録日 | 2008-09-16 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
4FC2
 
 | |
8XSF
 
 | | SARS-CoV-2 RBD + IMCAS-364 + hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, IMCAS-364 H chain, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.16 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSI
 
 | | SARS-CoV-2 RBD + IMCAS-364 (Local Refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-364 H chain, IMCAS-364 L chain, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSE
 
 | | SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 H chain, IMCAS-123 L chain, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8Y0Y
 
 | | Cryo-EM structure of the 123-316 scDb/PT-RBD complex | | 分子名称: | 123-316 scDb, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Jia, G.W, Tong, Z, Tong, J.Y, Su, Z.M. | | 登録日 | 2024-01-23 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSJ
 
 | | SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSL
 
 | | SARS-CoV-2 spike + IMCAS-123 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 heavy chain, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
5GNF
 
 | |
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | 分子名称: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | 著者 | Zhang, X, Ma, J, Wang, Y, Wang, J. | | 登録日 | 2016-08-07 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
5WIH
 
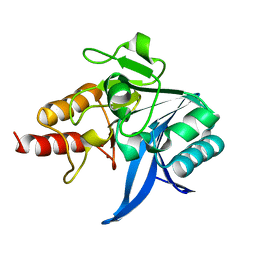 | |
5WIG
 
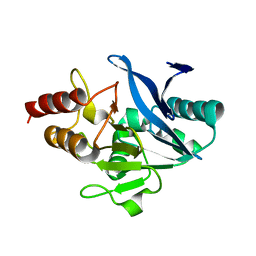 | |
4NA4
 
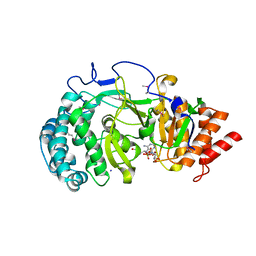 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADP-HPD | | 分子名称: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, IODIDE ION, Poly(ADP-ribose) glycohydrolase | | 著者 | Wang, Z, Cheng, Z, Xu, W. | | 登録日 | 2013-10-21 | | 公開日 | 2014-01-29 | | 最終更新日 | 2014-09-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4N9Y
 
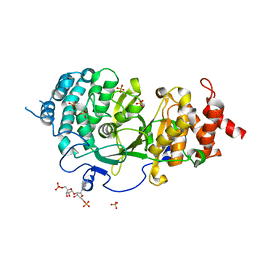 | |
4NA0
 
 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADPRibose | | 分子名称: | IODIDE ION, Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Wang, Z, Cheng, Z, Xu, W. | | 登録日 | 2013-10-21 | | 公開日 | 2014-01-29 | | 最終更新日 | 2014-09-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4N9Z
 
 | |
4NA5
 
 | |
4NA6
 
 | |
4MY2
 
 | | Crystal Structure of Norrin in fusion with Maltose Binding Protein | | 分子名称: | Maltose-binding periplasmic protein, Norrin fusion protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Ke, J, Jurecky, C, Chen, C, Gu, X, Parker, N, Williams, B.O, Melcher, K, Xu, H.E. | | 登録日 | 2013-09-27 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure and function of Norrin in assembly and activation of a Frizzled 4-Lrp5/6 complex.
Genes Dev., 27, 2013
|
|
