1XDP
 
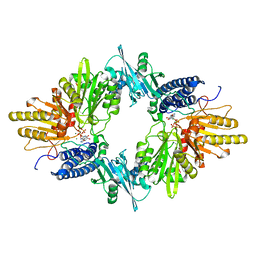 | | Crystal Structure of the E.coli Polyphosphate Kinase in complex with AMPPNP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyphosphate kinase | | 著者 | Zhu, Y, Huang, W, Lee, S.S, Xu, W. | | 登録日 | 2004-09-07 | | 公開日 | 2005-06-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis
Embo Rep., 6, 2005
|
|
1Z87
 
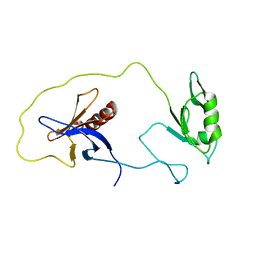 | | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | | 分子名称: | Alpha-1-syntrophin | | 著者 | Yan, J, Xu, W, Wen, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | 登録日 | 2005-03-30 | | 公開日 | 2006-01-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
1Z86
 
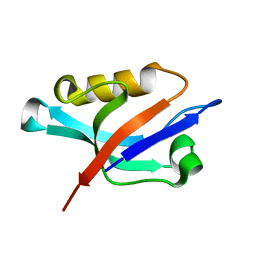 | | Solution structure of the PDZ domain of alpha-syntrophin | | 分子名称: | Alpha-1-syntrophin | | 著者 | Yan, J, Xu, W, Wen, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | 登録日 | 2005-03-30 | | 公開日 | 2006-01-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
1JDH
 
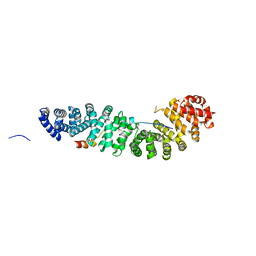 | | CRYSTAL STRUCTURE OF BETA-CATENIN AND HTCF-4 | | 分子名称: | BETA-CATENIN, hTcf-4 | | 著者 | Graham, T.A, Ferkey, D.M, Mao, F, Kimelman, D, Xu, W. | | 登録日 | 2001-06-13 | | 公開日 | 2001-12-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Tcf4 can specifically recognize beta-catenin using alternative conformations.
Nat.Struct.Biol., 8, 2001
|
|
1KBH
 
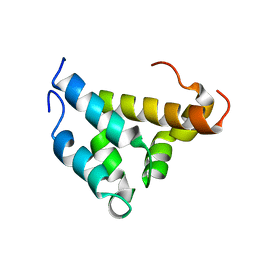 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | 分子名称: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | 著者 | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | 登録日 | 2001-11-06 | | 公開日 | 2002-02-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
1LUJ
 
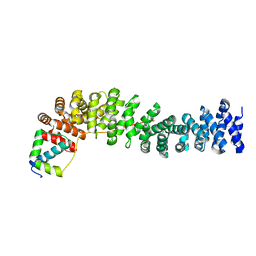 | | Crystal Structure of the Beta-catenin/ICAT Complex | | 分子名称: | Beta-catenin-interacting protein 1, Catenin beta-1 | | 著者 | Graham, T.A, Clements, W.K, Kimelman, D, Xu, W. | | 登録日 | 2002-05-22 | | 公開日 | 2002-10-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of the beta-catenin/ICAT complex reveals the inhibitory mechanism of ICAT.
Mol.Cell, 10, 2002
|
|
5EJC
 
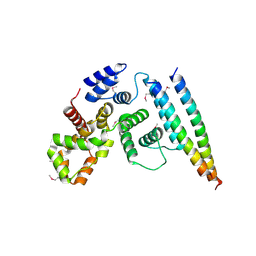 | | Crystal structural of the TSC1-TBC1D7 complex | | 分子名称: | Hamartin, TBC1 domain family member 7 | | 著者 | Wang, Z, Qin, J, Gong, W, Xu, W. | | 登録日 | 2015-11-01 | | 公開日 | 2016-03-02 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis of the Interaction between Tuberous Sclerosis Complex 1 (TSC1) and Tre2-Bub2-Cdc16 Domain Family Member 7 (TBC1D7).
J.Biol.Chem., 291, 2016
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
7MSY
 
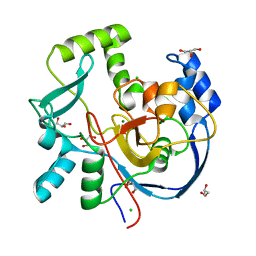 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | 分子名称: | CALCIUM ION, CHLORIDE ION, CalU17, ... | | 著者 | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2021-05-12 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7ML6
 
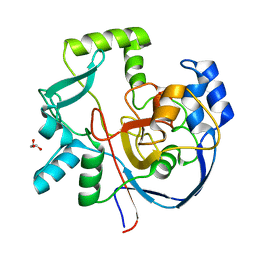 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | 分子名称: | CalU17, GLYCEROL | | 著者 | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2021-04-27 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
5HKP
 
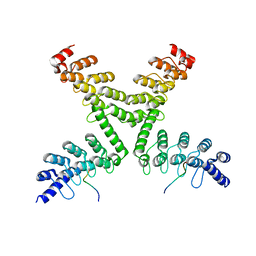 | | Crystal structure of mouse Tankyrase/human TRF1 complex | | 分子名称: | Tankyrase-1, Telomeric repeat-binding factor 1 | | 著者 | Wang, Z, Li, B, Rao, Z, Xu, W. | | 登録日 | 2016-01-14 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a tankyrase 1-telomere repeat factor 1 complex.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5UFL
 
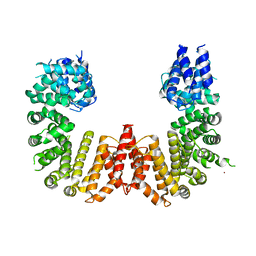 | | Crystal structure of a CIP2A core domain | | 分子名称: | Protein CIP2A, ZINC ION | | 著者 | Wang, Z, Wang, J, Rao, Z, Xu, W. | | 登録日 | 2017-01-04 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Oncoprotein CIP2A is stabilized via interaction with tumor suppressor PP2A/B56.
EMBO Rep., 18, 2017
|
|
3UTM
 
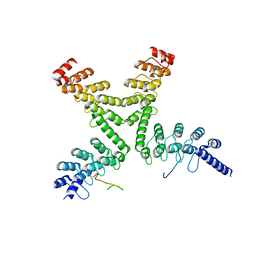 | |
3V3L
 
 | |
4ZAH
 
 | | Crystal structure of sugar aminotransferase WecE with External Aldimine VII from Escherichia coli K-12 | | 分子名称: | [[(2R,3S,5R)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R,4S,5R,6R)-6-methyl-5-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3,4-bis(oxidanyl)oxan-2-yl] hydrogen phosphate, dTDP-4-amino-4,6-dideoxygalactose transaminase | | 著者 | Wang, F, Singh, S, Cao, H, Xu, W, Miller, M.D, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural Basis for the Stereochemical Control of Amine Installation in Nucleotide Sugar Aminotransferases.
Acs Chem.Biol., 10, 2015
|
|
1QZ7
 
 | | Beta-catenin binding domain of Axin in complex with beta-catenin | | 分子名称: | Axin, Beta-catenin | | 著者 | Xing, Y, Clements, W.K, Kimelman, D, Xu, W. | | 登録日 | 2003-09-15 | | 公開日 | 2003-11-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a beta-catenin/Axin complex suggests a mechanism for the {beta}-catenin destruction complex
GENES DEV., 17, 2003
|
|
1TH1
 
 | | Beta-catenin in complex with a phosphorylated APC 20aa repeat fragment | | 分子名称: | Adenomatous polyposis coli protein, Beta-catenin | | 著者 | Xing, Y, Clements, W.K, Le Trong, I, Hinds, T.R, Stenkamp, R, Kimelman, D, Xu, W. | | 登録日 | 2004-05-31 | | 公開日 | 2004-09-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of a beta-Catenin/APC Complex Reveals a Critical Role for APC Phosphorylation in APC Function.
Mol.Cell, 15, 2004
|
|
2PF4
 
 | |
4EFE
 
 | | crystal structure of DNA ligase | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | 著者 | Wei, Y, Wang, T, Charifson, P, Xu, W. | | 登録日 | 2012-03-29 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | crystal structure of DNA ligase
To be Published
|
|
4EFB
 
 | | Crystal structure of DNA ligase | | 分子名称: | 4-amino-2-(cyclopentyloxy)-6-{[(1R,2S)-2-hydroxycyclopentyl]oxy}pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | 著者 | Wei, Y, Wang, T, Charifson, P, Xu, W. | | 登録日 | 2012-03-29 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of DNA ligase
To be Published
|
|
4FC2
 
 | |
8WUL
 
 | | Crystal structure of affinity enhanced TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | 分子名称: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen, ... | | 著者 | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | 登録日 | 2023-10-20 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
8WTE
 
 | | Crystal structure of TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | 分子名称: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen (Fragment), ... | | 著者 | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | 登録日 | 2023-10-18 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
