7X1L
 
 | | Malate dehydrogenase from Geobacillus stearothermophilus (gs-MDH) delta E311 mutant complexed with Nicotinamide Adenine Dinucleotide (NAD+) | | 分子名称: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | 登録日 | 2022-02-24 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Reducing substrate inhibition of malate dehydrogenase from Geobacillus stearothermophilus by C-terminal truncation.
Protein Eng.Des.Sel., 35, 2022
|
|
8ITH
 
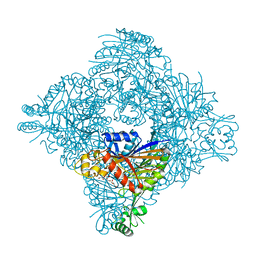 | |
8ITG
 
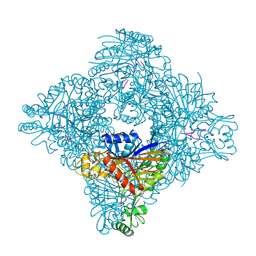 | |
5XOV
 
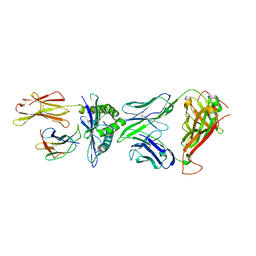 | | Crystal structure of peptide-HLA-A24 bound to S19-2 V-delta/V-beta TCR | | 分子名称: | Beta-2-microglobulin, HIV-1 Nef138-10 peptide, HLA class I histocompatibility antigen, ... | | 著者 | Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2017-05-31 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.684 Å) | | 主引用文献 | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
5XOS
 
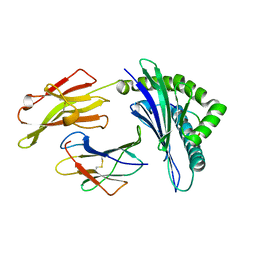 | | Crystal structure of HLA-B35 in complex with a pepetide antigen | | 分子名称: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2017-05-31 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.697 Å) | | 主引用文献 | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
5XOT
 
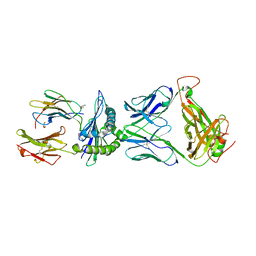 | | Crystal structure of pHLA-B35 in complex with TU55 T cell receptor | | 分子名称: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2017-05-31 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.787 Å) | | 主引用文献 | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
7BY9
 
 | | Malate Dehydrogenase from Geobacillus stearothermophilus (gs-MDH) complexed with Oxaloacetic Acid (OAA) and Nicotinamide Adenine Dinucleotide (NAD) | | 分子名称: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION | | 著者 | Shimozawa, Y, Nakamura, T, Himiyama, T, Nishiya, Y. | | 登録日 | 2020-04-22 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural analysis and reaction mechanism of malate dehydrogenase from Geobacillus stearothermophilus.
J.Biochem., 170, 2021
|
|
7BY8
 
 | |
7BYA
 
 | | Malate Dehydrogenase from Geobacillus stearothermophilus (gs-MDH) complexed with Oxaloacetic Acid (OAA) and Adenosine 5'-Diphosphoribose (APR) | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, Malate dehydrogenase, OXALOACETATE ION | | 著者 | Shimozawa, Y, Nakamura, T, Himiyama, T, Nishiya, Y. | | 登録日 | 2020-04-22 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural analysis and reaction mechanism of malate dehydrogenase from Geobacillus stearothermophilus.
J.Biochem., 170, 2021
|
|
4YN5
 
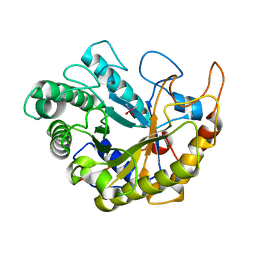 | | Catalytic domain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase | | 分子名称: | CACODYLATE ION, Mannan endo-1,4-beta-mannosidase | | 著者 | Shimane, Y, Ohta, Y, Usami, R, Hatada, Y. | | 登録日 | 2015-03-09 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase
To Be Published
|
|
6KV0
 
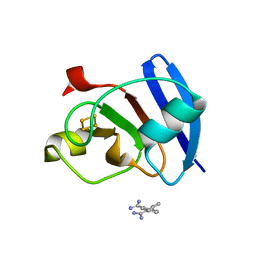 | | Ferredoxin I from C. reinhardtii, high X-ray dose | | 分子名称: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Onishi, Y, Kurisu, G, Tanaka, H. | | 登録日 | 2019-09-03 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6KUM
 
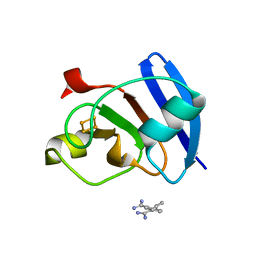 | | Ferredoxin I from C. reinhardtii, low X-ray dose | | 分子名称: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Onishi, Y, Kurisu, G, Tanaka, H. | | 登録日 | 2019-09-02 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6LK1
 
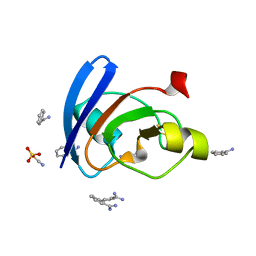 | | Ultrahigh resolution X-ray structure of Ferredoxin I from C. reinhardtii | | 分子名称: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | 著者 | Onishi, Y, Kurisu, G, Tanaka, H. | | 登録日 | 2019-12-17 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
8J6V
 
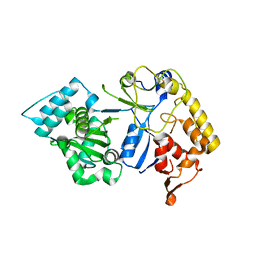 | |
6KS2
 
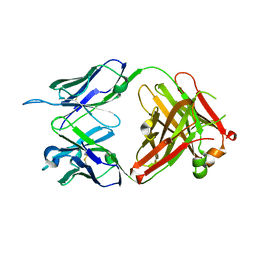 | | Structure of anti-Ghrelin receptor antibody | | 分子名称: | Fab7881 Heavy Chain (FabH), Fab7881 Light Chain (FabL) | | 著者 | Shiimura, Y, Horita, S, Asada, H, Hirata, K, Iwata, S, Kojima, M. | | 登録日 | 2019-08-23 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.753 Å) | | 主引用文献 | Structure of an antagonist-bound ghrelin receptor reveals possible ghrelin recognition mode.
Nat Commun, 11, 2020
|
|
6KO5
 
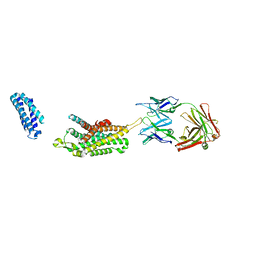 | | Complex structure of Ghrelin receptor with Fab | | 分子名称: | 6-(4-bromanyl-2-fluoranyl-phenoxy)-2-methyl-3-[[(3~{S})-1-propan-2-ylpiperidin-3-yl]methyl]pyrido[3,2-d]pyrimidin-4-one, Chimera of Soluble cytochrome b562 and Growth hormone secretagogue receptor type 1, Fab7881 Heavy Chain, ... | | 著者 | Shiimura, Y, Horita, S, Asada, H, Hirata, K, Iwata, S, Kojima, M. | | 登録日 | 2019-08-08 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of an antagonist-bound ghrelin receptor reveals possible ghrelin recognition mode.
Nat Commun, 11, 2020
|
|
7BT2
 
 | | Crystal structure of the SERCA2a in the E2.ATP state | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Kabashima, Y, Ogawa, H, Nakajima, R, Toyoshima, C. | | 登録日 | 2020-03-31 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.00002861 Å) | | 主引用文献 | What ATP binding does to the Ca2+pump and how nonproductive phosphoryl transfer is prevented in the absence of Ca2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8ZYO
 
 | | Cryo-EM Structure of astemizole-bound hERG Channel | | 分子名称: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Potassium voltage-gated channel subfamily H member 2 | | 著者 | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | 登録日 | 2024-06-18 | | 公開日 | 2024-09-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8ZYP
 
 | | Cryo-EM Structure of E-4031-bound hERG Channel | | 分子名称: | Potassium voltage-gated channel subfamily H member 2, ~{N}-[4-[1-[2-(6-methylpyridin-2-yl)ethyl]piperidin-4-yl]carbonylphenyl]methanesulfonamide | | 著者 | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | 登録日 | 2024-06-18 | | 公開日 | 2024-09-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8ZYN
 
 | | Cryo-EM Structure of inhibitor-free hERG Channel | | 分子名称: | Potassium voltage-gated channel subfamily H member 2 | | 著者 | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | 登録日 | 2024-06-18 | | 公開日 | 2024-09-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8ZYQ
 
 | | Cryo-EM Structure of pimozide-bound hERG Channel | | 分子名称: | 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, Potassium voltage-gated channel subfamily H member 2 | | 著者 | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | 登録日 | 2024-06-18 | | 公開日 | 2024-09-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
1BYV
 
 | | GLYCOSYLATED EEL CALCITONIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CALCITONIN) | | 著者 | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K.G, Opella, S.J. | | 登録日 | 1998-10-16 | | 公開日 | 1998-10-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
1BZB
 
 | | GLYCOSYLATED EEL CALCITONIN | | 分子名称: | PROTEIN (CALCITONIN), alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | 登録日 | 1998-10-27 | | 公開日 | 1998-11-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
1BKU
 
 | | EFFECTS OF GLYCOSYLATION ON THE STRUCTURE AND DYNAMICS OF EEL CALCITONIN, NMR, 10 STRUCTURES | | 分子名称: | CALCITONIN | | 著者 | Hashimoto, Y, Nishikido, J, Toma, K, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | 登録日 | 1998-07-13 | | 公開日 | 1999-01-13 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
6T1H
 
 | | OXA-51-like beta-lactamase OXA-66 | | 分子名称: | Beta-lactamase OXA-66, ZINC ION | | 著者 | Takebayashi, Y, Chirgadze, D, Henderson, S, Warburton, P.J, Evans, B.E. | | 登録日 | 2019-10-04 | | 公開日 | 2020-10-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the OXA-51-like beta-lactamase OXA-66
To Be Published
|
|
