6LUK
 
 | |
6LUI
 
 | | Crystal structure of the SAMD1 WH domain and DNA complex | | 分子名称: | Atherin, DNA (5'-D(*AP*CP*CP*TP*GP*CP*GP*CP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*AP*TP*GP*GP*TP*GP*CP*GP*CP*AP*GP*GP*T)-3') | | 著者 | Zhou, Y, Cao, Y, Wang, Z. | | 登録日 | 2020-01-29 | | 公開日 | 2021-02-03 | | 最終更新日 | 2021-07-07 | | 実験手法 | X-RAY DIFFRACTION (1.781 Å) | | 主引用文献 | The SAM domain-containing protein 1 (SAMD1) acts as a repressive chromatin regulator at unmethylated CpG islands.
Sci Adv, 7, 2021
|
|
6LUJ
 
 | |
4L07
 
 | | Crystal structure of the maleamate amidase Ami from Pseudomonas putida S16 | | 分子名称: | GLYCEROL, Hydrolase, isochorismatase family, ... | | 著者 | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | 登録日 | 2013-05-30 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
4L08
 
 | | Crystal structure of the maleamate amidase Ami(C149A) in complex with maleate from Pseudomonas putida S16 | | 分子名称: | Hydrolase, isochorismatase family, MALEIC ACID | | 著者 | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | 登録日 | 2013-05-31 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
4K7F
 
 | | Newly identified epitope V60 from HBV core protein complexed with HLA-A*0201 | | 分子名称: | Beta-2-microglobulin, Core protein, HLA class I histocompatibility antigen, ... | | 著者 | Meng, S.D, Zhang, Y, Wu, Y, Qi, J.X. | | 登録日 | 2013-04-17 | | 公開日 | 2013-06-05 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The L60V variation in hepatitis B virus core protein elicits new epitope-specific cytotoxic T lymphocytes and enhances viral replication.
J.Virol., 87, 2013
|
|
8J8Q
 
 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | 分子名称: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | 著者 | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | 登録日 | 2023-05-02 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
8J8P
 
 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | 分子名称: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | 著者 | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | 登録日 | 2023-05-02 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
8JJO
 
 | | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with an antagonist | | 分子名称: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, Beta-2 adrenergic receptor,Beta-2 adrenergic receptor,Soluble cytochrome b562 | | 著者 | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | 登録日 | 2023-05-31 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8J7E
 
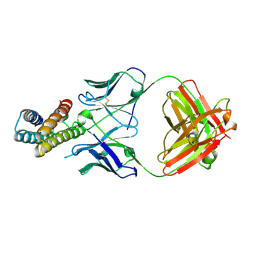 | |
8JJL
 
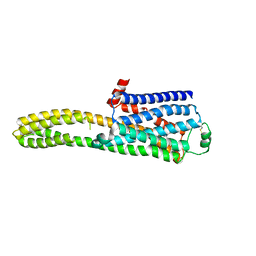 | | cryo-EM structure of the beta2-AR-mBRIL/1b3 Fab/Glue complex with a full agonist | | 分子名称: | Beta-2 adrenergic receptor,Soluble cytochrome b562, Olodaterol | | 著者 | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | 登録日 | 2023-05-30 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JJ8
 
 | | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with a partial agonist | | 分子名称: | Beta-2 adrenergic receptor,Soluble cytochrome b562, ~{N}-[5-[(1~{R})-2-[[(2~{R})-1-(4-methoxyphenyl)propan-2-yl]amino]-1-oxidanyl-ethyl]-2-oxidanyl-phenyl]methanamide | | 著者 | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | 登録日 | 2023-05-29 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JMT
 
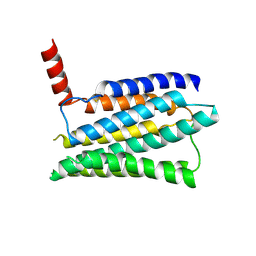 | | Structure of the adhesion GPCR ADGRL3 in the apo state | | 分子名称: | Adhesion G protein-coupled receptor L3,Soluble cytochrome b562 | | 著者 | Tao, Y, Guo, Q, He, B, Zhong, Y. | | 登録日 | 2023-06-05 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8IP5
 
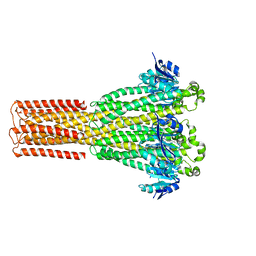 | | Cryo-EM structure of hMRS2-lowEDTA | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | 著者 | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | 登録日 | 2023-03-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP3
 
 | | Cryo-EM structure of hMRS2-Mg | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | 著者 | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | 登録日 | 2023-03-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP6
 
 | | Cryo-EM structure of hMRS2-rest | | 分子名称: | CHLORIDE ION, Magnesium transporter MRS2 homolog, mitochondrial | | 著者 | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | 登録日 | 2023-03-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP4
 
 | | Cryo-EM structure of hMRS-highEDTA | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | 著者 | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | 登録日 | 2023-03-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8JBA
 
 | |
8H7A
 
 | |
8K0D
 
 | |
8K0C
 
 | |
7X2K
 
 | | Crystal structure of nanobody Nb70 with antibody 1F11 fab and SARS-CoV-2 RBD | | 分子名称: | 1F11-H, 1F11-L, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2J
 
 | | Crystal structure of nanobody Nb70 with SARS-CoV RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb70, Spike protein S1 | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | 分子名称: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
3IKI
 
 | | 5-SMe-dU containing DNA octamer | | 分子名称: | 5'-D(*GP*(UMS)P*GP*(US2)P*AP*CP*AP*C)-3', MAGNESIUM ION | | 著者 | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | 登録日 | 2009-08-05 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Nucleic Acids Res., 40, 2012
|
|
