2KAF
 
 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | 分子名称: | Non-structural protein 3 | | 著者 | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-11-05 | | 公開日 | 2008-11-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
1QFU
 
 | | INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH A NEUTRALIZING ANTIBODY | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (HEMAGGLUTININ (HA1 CHAIN)), ... | | 著者 | Fleury, D, Gigant, B, Bizebard, T, Daniels, R.S, Skehel, J.J, Knossow, M. | | 登録日 | 1999-04-14 | | 公開日 | 1999-04-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site.
Nat.Struct.Biol., 6, 1999
|
|
1QA0
 
 | | BOVINE TRYPSIN 2-AMINOBENZIMIDAZOLE COMPLEX | | 分子名称: | 2H-BENZOIMIDAZOL-2-YLAMINE, CALCIUM ION, TRYPSIN | | 著者 | Whitlow, M. | | 登録日 | 1999-04-09 | | 公開日 | 2000-04-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2KYS
 
 | | NMR Structure of the SARS Coronavirus Nonstructural Protein Nsp7 in Solution at pH 6.5 | | 分子名称: | Non-structural protein 7 | | 著者 | Johnson, M.A, Jaudzems, K, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2010-06-07 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the SARS-CoV Nonstructural Protein 7 in Solution at pH 6.5.
J.Mol.Biol., 402, 2010
|
|
6PZW
 
 | | CryoEM derived model of NA-22 Fab in complex with N9 Shanghai2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-22 fragment antigen binding heavy chain, ... | | 著者 | Ward, A.B, Turner, H.L, Zhu, X. | | 登録日 | 2019-08-01 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-04-05 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6PW6
 
 | | The HIV-1 Envelope Glycoprotein Clone BG505 SOSIP.664 in Complex with Three Copies of the Bovine Broadly Neutralizing Antibody, NC-Cow1, Fragment Antigen Binding Domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Broadly Neutralizing Antibody NC-Cow1 Heavy Chain, ... | | 著者 | Berndsen, Z.T, Ward, A.B. | | 登録日 | 2019-07-22 | | 公開日 | 2020-06-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural basis of broad HIV neutralization by a vaccine-induced cow antibody.
Sci Adv, 6, 2020
|
|
6PZZ
 
 | | CryoEM derived model of NA-80 Fab in complex with N9 Shanghai2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-80 fragment antibody heavy chain, ... | | 著者 | Ward, A.B, Turner, H.L, Zhu, X. | | 登録日 | 2019-08-01 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-04-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6PZY
 
 | | CryoEM derived model of NA-73 Fab in complex with N9 Shanghai2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-73 fragment antibody heavy chain, ... | | 著者 | Ward, A.B, Turner, H.L, Zhu, X. | | 登録日 | 2019-08-01 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-04-05 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
3YPI
 
 | |
4Q5K
 
 | | Crystal structure of a N-acetylmuramoyl-L-alanine amidase (BACUNI_02947) from Bacteroides uniformis ATCC 8492 at 1.30 A resolution | | 分子名称: | (2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoic acid, SODIUM ION, Uncharacterized protein | | 著者 | Joint Center for Structural Genomics (JCSG) | | 登録日 | 2014-04-17 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure-guided functional characterization of DUF1460 reveals a highly specific NlpC/P60 amidase family.
Structure, 22, 2014
|
|
4Q98
 
 | |
4QB7
 
 | |
4Q68
 
 | |
4RDB
 
 | |
4R8O
 
 | |
4R03
 
 | |
4RBO
 
 | |
4U6G
 
 | | Crystal structure of 10E8 Fab in complex with a hydrocarbon-stapled HIV-1 gp41 MPER peptide | | 分子名称: | 10E8 Fab Heavy Chain, 10E8 Fab Light Chain, SAH-MPER(662-683KKK)(B,q) | | 著者 | Ofek, G, Bird, G.H, Walensky, L.D, Kwong, P.D. | | 登録日 | 2014-07-28 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (4.2012 Å) | | 主引用文献 | Structural Fidelity and Neutralizing Antibody Binding Capacity of
Hydrocarbon-Stapled HIV-1 Antigens
To be published
|
|
6W4Y
 
 | | Structure of full-length human lambda-6A light chain JTO in complex with hydantoin stabilizer | | 分子名称: | 2-[(4~{R})-4-(2-methylpropyl)-2,5-bis(oxidanylidene)imidazolidin-1-yl]-~{N}-[4-(trifluoromethyl)phenyl]ethanamide, GLYCEROL, JTO light chain, ... | | 著者 | Yan, N.L, Morgan, G.J, Kelly, J.W. | | 登録日 | 2020-03-11 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural basis for the stabilization of amyloidogenic immunoglobulin light chains by hydantoins.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
2OOK
 
 | |
2OOC
 
 | |
2PV7
 
 | |
2Q3L
 
 | |
2Q86
 
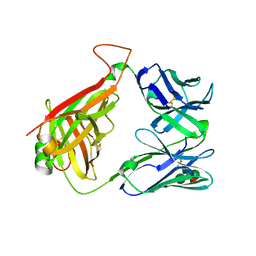 | | Structure of the mouse invariant NKT cell receptor Valpha14 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Valpha14 TCR, Vbeta8.2, ... | | 著者 | Zajonc, D.M. | | 登録日 | 2007-06-08 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structures of Mouse CD1d-iGb3 Complex and its Cognate Valpha14 T Cell Receptor Suggest a Model for Dual Recognition of Foreign and Self Glycolipids.
J.Mol.Biol., 377, 2008
|
|
2Q8U
 
 | |
