8J34
 
 | | Crystal structure of MERS main protease in complex with PF00835231 | | 分子名称: | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of MERS main protease in complex with PF00835231
To Be Published
|
|
8J3B
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF00835231
To Be Published
|
|
8J39
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF00835231
To Be Published
|
|
4EMP
 
 | | Crystal structure of the mutant of ClpP E137A from Staphylococcus aureus | | 分子名称: | ATP-dependent Clp protease proteolytic subunit | | 著者 | Ye, F, Zhang, J, Liu, H, Luo, C, Yang, C.-G. | | 登録日 | 2012-04-12 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease
J.Biol.Chem., 288, 2013
|
|
8J36
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
M49I mutant in complex with PF00835231
To Be Published
|
|
8JC6
 
 | |
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | 登録日 | 2020-06-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
5H56
 
 | | ADP and dTDP bound Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | 著者 | Biswas, A, Jeyakanthan, J, Sekar, K, Kuramitsu, S, Yokoyama, S. | | 登録日 | 2016-11-04 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural studies of a hyperthermophilic thymidylate kinase enzyme reveal conformational substates along the reaction coordinate
FEBS J., 284, 2017
|
|
2Z6Y
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | 分子名称: | Fluorescent protein Dronpa | | 著者 | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | 登録日 | 2007-08-09 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z6Z
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | 分子名称: | Fluorescent protein Dronpa | | 著者 | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | 登録日 | 2007-08-09 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5Z6P
 
 | | The crystal structure of an agarase, AgWH50C | | 分子名称: | B-agarase | | 著者 | Mao, X, Zhou, J, Zhang, P, Zhang, L, Zhang, J, Li, Y. | | 登録日 | 2018-01-24 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.061 Å) | | 主引用文献 | Structure-based design of agarase AgWH50C from Agarivorans gilvus WH0801 to enhance thermostability.
Appl. Microbiol. Biotechnol., 103, 2019
|
|
3P20
 
 | | Crystal structure of vanadate bound subunit A of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | 著者 | Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | 登録日 | 2010-10-01 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | The transition-like state and Pi entrance into the catalytic a subunit of the biological engine A-ATP synthase.
J.Mol.Biol., 408, 2011
|
|
2LI6
 
 | |
3CIS
 
 | | The Crystal Structure of Rv2623 from Mycobacterium tuberculosis | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein | | 著者 | Bilder, P, Drumm, J, Mi, K, Chan, J, Almo, S.C. | | 登録日 | 2008-03-11 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Crystal Structure of Rv2623 : a Novel, Tandem-Repeat Universal Stress Protein of Mycobacterium tuberculosis
To be Published
|
|
2L6K
 
 | | Solution Structure of a Nonphosphorylated Peptide Recognizing Domain | | 分子名称: | Tensin-like C1 domain-containing phosphatase | | 著者 | Dai, K, Liao, S, Zhang, J, Zhang, X, Tu, X. | | 登録日 | 2010-11-22 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Tensin2 SH2 domain and its phosphotyrosine-independent interaction with DLC-1
Plos One, 6, 2011
|
|
2LNX
 
 | |
1X3E
 
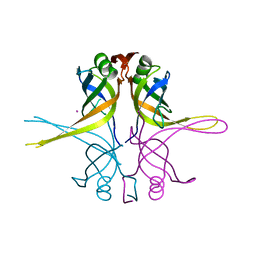 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium smegmatis | | 分子名称: | CADMIUM ION, Single-strand binding protein | | 著者 | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | 登録日 | 2005-05-04 | | 公開日 | 2005-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ABV
 
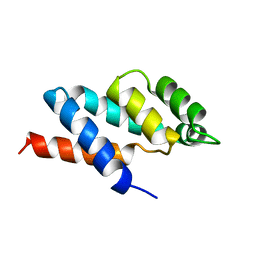 | | N-TERMINAL DOMAIN OF THE DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE FROM ESCHERICHIA COLI, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE | | 著者 | Wilkens, S, Dunn, S.D, Chandler, J, Dahlquist, F.W, Capaldi, R.A. | | 登録日 | 1997-01-29 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal domain of the delta subunit of the E. coli ATPsynthase.
Nat.Struct.Biol., 4, 1997
|
|
3V7Q
 
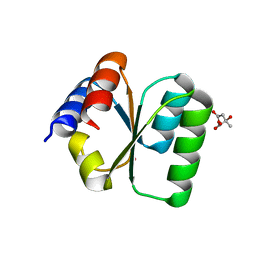 | | Crystal structure of B. subtilis YlxQ at 1.55 A resolution | | 分子名称: | CITRIC ACID, POTASSIUM ION, Probable ribosomal protein ylxQ | | 著者 | Baird, N.J, Zhang, J, Hamma, T, Ferre-D'Amare, A.R. | | 登録日 | 2011-12-21 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops.
Rna, 18, 2012
|
|
1X3G
 
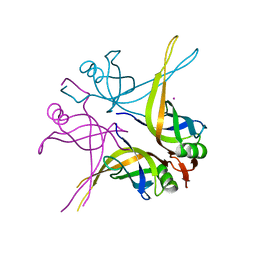 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | 分子名称: | CADMIUM ION, Single-strand binding protein | | 著者 | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | 登録日 | 2005-05-05 | | 公開日 | 2005-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | 著者 | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | 登録日 | 2020-11-22 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.199 Å) | | 主引用文献 | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
7DJZ
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW01 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, MW01 heavy chain, ... | | 著者 | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | 登録日 | 2020-11-22 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.397 Å) | | 主引用文献 | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
1X3F
 
 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | 分子名称: | CADMIUM ION, Single-strand binding protein | | 著者 | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | 登録日 | 2005-05-05 | | 公開日 | 2005-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2ID0
 
 | | Escherichia coli RNase II | | 分子名称: | Exoribonuclease 2, MANGANESE (II) ION | | 著者 | Zuo, Y, Zhang, J, Wang, Y, Malhotra, A. | | 登録日 | 2006-09-13 | | 公開日 | 2006-10-03 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural Basis for Processivity and Single-Strand Specificity of RNase II.
Mol.Cell, 24, 2006
|
|
5Y4M
 
 | | Discoidin domain of human CASPR2 | | 分子名称: | 1,2-ETHANEDIOL, human CASPR2 Disc domain | | 著者 | Liu, H, Xu, F, Zhang, J, Liang, W. | | 登録日 | 2017-08-04 | | 公開日 | 2018-08-08 | | 最終更新日 | 2019-02-20 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Structural mapping of hot spots within human CASPR2 discoidin domain for autoantibody recognition.
J. Autoimmun., 96, 2019
|
|
