7WH8
 
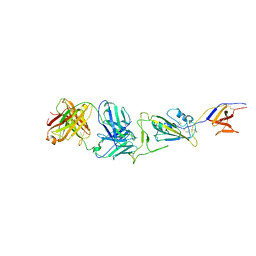 | |
7WHB
 
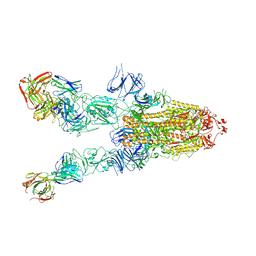 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Zeng, J.W, Ge, J.W, Wang, X.Q. | | 登録日 | 2021-12-30 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (2.67 Å) | | 主引用文献 | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
7VK9
 
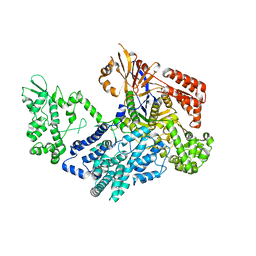 | | Crystal structure of xCas9 P411T | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1 | | 著者 | Bao, R, Liu, H.Y, Luo, Y.Z, Song, Y.J. | | 登録日 | 2021-09-29 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and Dynamics Studies of the Spcas9 Variant Provide Insights into the Regulatory Role of the REC1 Domain
Acs Catalysis, 12, 2022
|
|
7VAH
 
 | |
7WHD
 
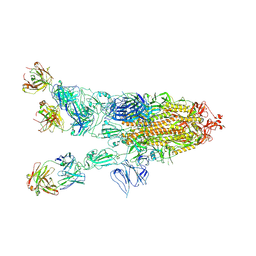 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Zeng, J.W, Wang, X.W, Ge, J.W, Wang, Z.Y. | | 登録日 | 2021-12-30 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
6J66
 
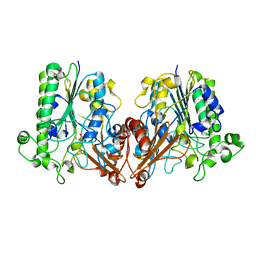 | | Chondroitin sulfate/dermatan sulfate endolytic 4-O-sulfatase | | 分子名称: | CALCIUM ION, Chondroitin sulfate/dermatan sulfate 4-O-endosulfatase protein | | 著者 | Gu, L, Li, F, Su, T, Wang, S. | | 登録日 | 2019-01-14 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.953 Å) | | 主引用文献 | Comparative Study of Two Chondroitin Sulfate/Dermatan Sulfate 4-O-Sulfatases With High Identity.
Front Microbiol, 10, 2019
|
|
6JYS
 
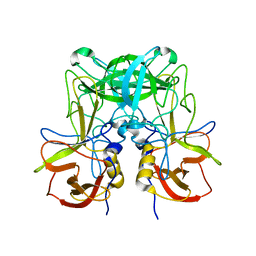 | |
5YKR
 
 | |
5YKT
 
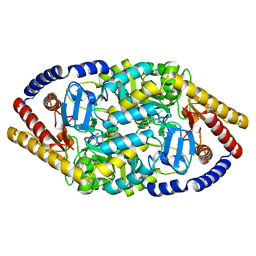 | |
6JYO
 
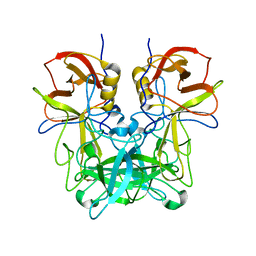 | |
6K2O
 
 | |
6K2N
 
 | |
6JYN
 
 | |
6JYR
 
 | |
7EPM
 
 | | human LDHC complexed with NAD+ and ethylamino acetic acid | | 分子名称: | 2-(ethylamino)-2-oxidanylidene-ethanoic acid, L-lactate dehydrogenase C chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Yu, Y, Chen, Q. | | 登録日 | 2021-04-27 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Identification of human LDHC4 as a potential target for anticancer drug discovery.
Acta Pharm Sin B, 12, 2022
|
|
2L1V
 
 | |
2JVA
 
 | | NMR solution structure of peptidyl-tRNA hydrolase domain protein from Pseudomonas syringae pv. tomato. Northeast Structural Genomics Consortium target PsR211 | | 分子名称: | Peptidyl-tRNA hydrolase domain protein | | 著者 | Singarapu, K.K, Sukumaran, D, Parish, D, Eletsky, A, Zhang, Q, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-09-14 | | 公開日 | 2007-10-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors.
Proteins, 71, 2008
|
|
7EAX
 
 | | Crystal complex of p53-V272M and antimony ion | | 分子名称: | ANTIMONY (III) ION, Cellular tumor antigen p53, ZINC ION | | 著者 | Lu, M, Tang, Y. | | 登録日 | 2021-03-08 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Repurposing antiparasitic antimonials to noncovalently rescue temperature-sensitive p53 mutations.
Cell Rep, 39, 2022
|
|
7E5X
 
 | |
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp4/5 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-14 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp14/15 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp15/16 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp9/10 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp6/7 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
