8IW7
 
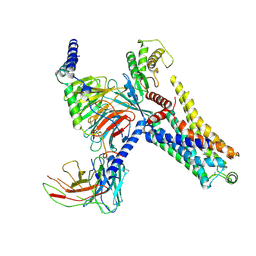 | | Cryo-EM structure of the PEA-bound mTAAR9-Gs complex | | 分子名称: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWE
 
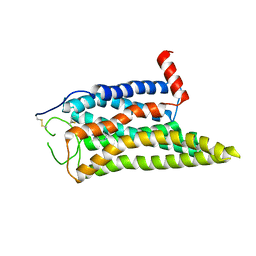 | | Cryo-EM structure of the SPE-mTAAR9 complex | | 分子名称: | SPERMIDINE, Trace amine-associated receptor 9 | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWM
 
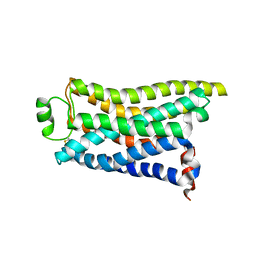 | | Cryo-EM structure of the PEA-bound mTAAR9 complex | | 分子名称: | 2-PHENYLETHYLAMINE, Trace amine-associated receptor 9 | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-30 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW1
 
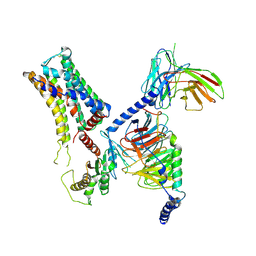 | | Cryo-EM structure of the PEA-bound mTAAR9-Golf complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(olf) subunit alpha, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
6UYM
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2mc3-v6 redesigned core from genotype 1a bound to broadly neutralizing antibody AR3C | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | 著者 | Tzarum, N, Wilson, I.A, Zhu, J. | | 登録日 | 2019-11-13 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.848 Å) | | 主引用文献 | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
6UYF
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2mc3-v1 redesigned core from genotype 6a bound to broadly neutralizing antibody AR3B | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | 著者 | Tzarum, N, Wilson, I.A, Zhu, J. | | 登録日 | 2019-11-13 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
7DLZ
 
 | | Crystal Structure of Methyltransferase Ribozyme | | 分子名称: | RNA (45-MER), U1 small nuclear ribonucleoprotein A | | 著者 | Gan, J.H, Gao, Y.Q, Jiang, H.Y, Chen, D.R, Murchie, A.I.H. | | 登録日 | 2020-11-30 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.002 Å) | | 主引用文献 | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
6UYG
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2c3 core from genotype 6a bound to broadly neutralizing antibody AR3A and non neutralizing antibody E1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | 著者 | Tzarum, N, Wilson, I.A, Zhu, J. | | 登録日 | 2019-11-13 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.375 Å) | | 主引用文献 | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
6VIZ
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 27 | | 分子名称: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | 著者 | Longenecker, K.L, Park, C.H, Qiu, W. | | 登録日 | 2020-01-14 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.391 Å) | | 主引用文献 | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6VIW
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 18 | | 分子名称: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | 著者 | Longenecker, K.L, Park, C.H, Qiu, W. | | 登録日 | 2020-01-14 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.429 Å) | | 主引用文献 | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
8H77
 
 | | Hsp90-AhR-p23-XAP2 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | 著者 | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | 登録日 | 2022-10-19 | | 公開日 | 2023-01-04 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
6VIX
 
 | | BRD4_Bromodomain2 complex with pyrrolopyridone compound 18 | | 分子名称: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | 著者 | Longenecker, K.L, Park, C.H, Qiu, W. | | 登録日 | 2020-01-14 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.116 Å) | | 主引用文献 | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6VIY
 
 | | BRD2_Bromodomain2 complex with pyrrolopyridone compound 27 | | 分子名称: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 2 | | 著者 | Longenecker, K.L, Park, C.H, Qiu, W. | | 登録日 | 2020-01-14 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.904 Å) | | 主引用文献 | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
4H3F
 
 | | Structure of BACE Bound to 3-(5-((7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-3-yl)benzonitrile | | 分子名称: | 3-{5-[(2E,4aR,7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Mandal, M. | | 登録日 | 2012-09-13 | | 公開日 | 2012-11-07 | | 最終更新日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3G
 
 | | Structure of BACE Bound to 2-((7aR)-7a-(4-(3-cyanophenyl)thiophen-2-yl)-2-imino-3-methyl-4-oxohexahydro-1H-pyrrolo[3,4-d]pyrimidin-6(2H)-yl)nicotinonitrile | | 分子名称: | 2-{(2E,4aR,7aR)-7a-[4-(3-cyanophenyl)thiophen-2-yl]-2-imino-3-methyl-4-oxooctahydro-6H-pyrrolo[3,4-d]pyrimidin-6-yl}pyridine-3-carbonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Mandal, M. | | 登録日 | 2012-09-13 | | 公開日 | 2012-11-07 | | 最終更新日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3J
 
 | | Structure of BACE Bound to 2-fluoro-5-(5-(2-imino-3-methyl-4-oxo-6-phenyloctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-2-yl)benzonitrile | | 分子名称: | 2-fluoro-5-{5-[(2E,4aR,7aR)-2-imino-3-methyl-4-oxo-6-phenyloctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-2-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Mandal, M. | | 登録日 | 2012-09-13 | | 公開日 | 2012-10-17 | | 最終更新日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3I
 
 | | Structure of BACE Bound to 3-(5-((7aR)-2-imino-6-(3-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-3-yl)benzonitrile | | 分子名称: | 3-{5-[(2E,4aR,7aR)-2-imino-6-(3-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Mandal, M. | | 登録日 | 2012-09-13 | | 公開日 | 2012-11-07 | | 最終更新日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4HA5
 
 | | Structure of BACE Bound to (S)-3-(5-(2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl)thiophen-3-yl)benzonitrile | | 分子名称: | 3-{5-[(2E,4S)-2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Mandal, M. | | 登録日 | 2012-09-25 | | 公開日 | 2012-10-17 | | 最終更新日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
1QP9
 
 | | STRUCTURE OF HAP1-PC7 COMPLEXED TO THE UAS OF CYC7 | | 分子名称: | CYP1(HAP1-PC7) ACTIVATORY PROTEIN, DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), ... | | 著者 | Lukens, A, King, D, Marmorstein, R. | | 登録日 | 1999-06-01 | | 公開日 | 2000-10-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of HAP1-PC7 bound to DNA: implications for DNA recognition and allosteric effects of DNA-binding on transcriptional activation.
Nucleic Acids Res., 28, 2000
|
|
4L72
 
 | | Crystal structure of MERS-CoV complexed with human DPP4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | 著者 | Wang, X.Q, Wang, N.S. | | 登録日 | 2013-06-13 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.005 Å) | | 主引用文献 | Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4
Cell Res., 23, 2013
|
|
8GRQ
 
 | | Cryo-EM structure of BRCA1/BARD1 bound to H2AK127-UbcH5c-Ub nucleosome | | 分子名称: | BRCA1-associated RING domain protein 1, Breast cancer type 1 susceptibility protein, DNA (147-MER), ... | | 著者 | Ai, H.S, Zebin, T, Zhiheng, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | 登録日 | 2022-09-02 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
8GZ8
 
 | |
8GZ9
 
 | |
8DQA
 
 | |
8DQ6
 
 | |
