6JN3
 
 | |
5JW0
 
 | | Crystal structure of mithramycin analogue MTM SA-Phe in complex with a 10-mer DNA AGGGTACCCT | | 分子名称: | DNA (5'-D(P*AP*GP*GP*GP*TP*AP*CP*CP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Phe, ... | | 著者 | Hou, C, Rohr, J, Tsodikov, O.V. | | 登録日 | 2016-05-11 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
7X45
 
 | | Grass carp interferon gamma related | | 分子名称: | Interferon gamma | | 著者 | Wang, J, Zou, J, Zhu, X. | | 登録日 | 2022-03-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-04-19 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Novel Dimeric Architecture of an IFN-gamma-Related Cytokine Provides Insights into Subfunctionalization of Type II IFNs in Teleost Fish.
J Immunol., 209, 2022
|
|
5NHH
 
 | | Human Erk2 with an Erk1/2 inhibitor | | 分子名称: | 5-(2-methoxyethyl)-2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | 著者 | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | 登録日 | 2017-03-21 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NGU
 
 | | Human Erk2 with an Erk1/2 inhibitor | | 分子名称: | 2-[2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | 著者 | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | 登録日 | 2017-03-20 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHO
 
 | | Human Erk2 with an Erk1/2 inhibitor | | 分子名称: | (6~{S})-5-(2-methoxyethyl)-6-methyl-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | 著者 | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | 登録日 | 2017-03-22 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHV
 
 | | Human Erk2 with an Erk1/2 inhibitor | | 分子名称: | 7-[2-(oxan-4-ylamino)pyrimidin-4-yl]-3,4-dihydro-2~{H}-pyrrolo[1,2-a]pyrazin-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | 著者 | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | 登録日 | 2017-03-22 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
6JN6
 
 | |
5OR1
 
 | | BamA structure of Salmonella enterica | | 分子名称: | Outer membrane protein assembly factor BamA | | 著者 | Dong, C, Gu, Y. | | 登録日 | 2017-08-14 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | BamA beta 16C strand and periplasmic turns are critical for outer membrane protein insertion and assembly.
Biochem. J., 474, 2017
|
|
5JW2
 
 | | Crystal structure of mithramycin analogue MTM SA-Phe in complex with a 10-mer DNA AGGGATCCCT | | 分子名称: | DNA (5'-D(*AP*GP*GP*GP*AP*TP*CP*CP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Phe, ... | | 著者 | Hou, C, Rohr, J, Tsodikov, O.V. | | 登録日 | 2016-05-11 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
5JVT
 
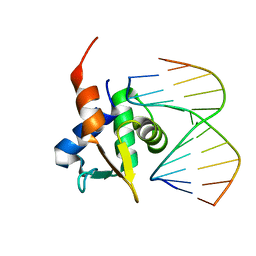 | |
7XMX
 
 | |
7XMZ
 
 | |
5K4Z
 
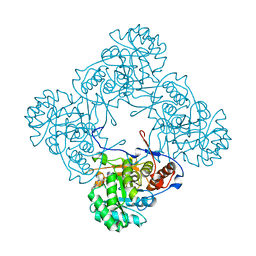 | | M. thermoresistible IMPDH in complex with IMP and Compound 6 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(4-fluorophenyl)-4-(2~{H}-indazol-6-ylsulfamoyl)-3,5-dimethyl-1~{H}-pyrrole-2-carboxamide | | 著者 | Pacitto, A, Ascher, D.B, Blundell, T.L. | | 登録日 | 2016-05-22 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
4ZQI
 
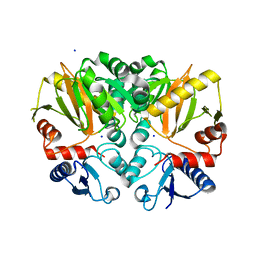 | | Crystal structure of Apo D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | 分子名称: | D-alanine--D-alanine ligase, SODIUM ION | | 著者 | Tran, H.-T, Kang, L.-W, Hong, M.-K, Ngo, H.P.T, Huynh, K.H, Ahn, Y.J. | | 登録日 | 2015-05-10 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | 分子名称: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | 著者 | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | 登録日 | 2020-07-29 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
5BPF
 
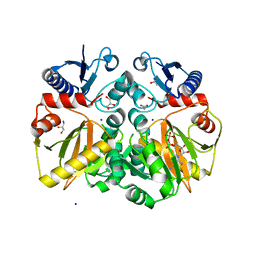 | | Crystal structure of ADP complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | 分子名称: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase, ... | | 著者 | Tran, H.T, Kang, L.W, Hong, M.K. | | 登録日 | 2015-05-28 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5BPH
 
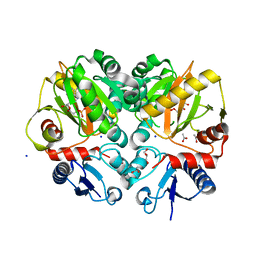 | | Crystal structure of AMP complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | 分子名称: | ACETATE ION, ADENOSINE MONOPHOSPHATE, D-alanine--D-alanine ligase, ... | | 著者 | Tran, H.T, Kang, L.W, Hong, M.K. | | 登録日 | 2015-05-28 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5B73
 
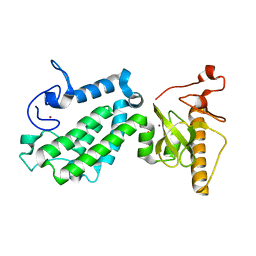 | |
3D1B
 
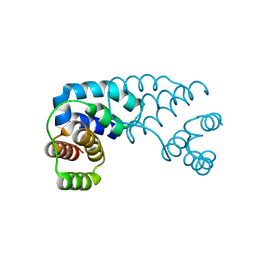 | |
5CD3
 
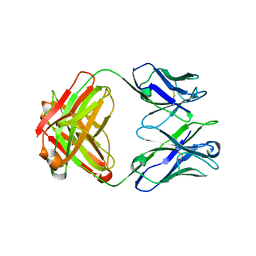 | |
5C1O
 
 | | Crystal structure of AMP-PNP complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | 分子名称: | D-alanine--D-alanine ligase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Tran, H.T, Kang, L.W, Hong, M.K. | | 登録日 | 2015-06-15 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7K6O
 
 | | Crystal structure of PI3Kalpha inhibitor 10-5429 | | 分子名称: | (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-N-methylpyrrolidine-1-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | 登録日 | 2020-09-21 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.738 Å) | | 主引用文献 | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
7K71
 
 | | Crystal structure of PI3Kalpha inhibitor 4-0686 | | 分子名称: | 2-(morpholin-4-yl)[4,5'-bipyrimidin]-2'-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | 登録日 | 2020-09-21 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
3D1D
 
 | |
