5ZAS
 
 | |
5AGR
 
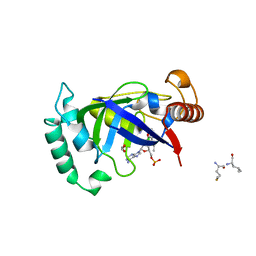 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | 分子名称: | 1,2-ETHANEDIOL, 3-AMINOMETHYL-7-(ETHOXY)-3H-BENZO[C][1,2]OXABOROL-1-OL modified adenosine, LEUCINE, ... | | 著者 | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | 登録日 | 2015-02-03 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
8V4P
 
 | |
8VEH
 
 | |
8V4O
 
 | |
8V5G
 
 | |
9AT3
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with an ethylcyclohexyl 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-{[N-({[(2S)-1-(2-cyclohexylethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-(2-cyclohexylethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9ASV
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a benzyl 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-{[N-({[(2S)-1-benzyl-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-benzyl-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9ASW
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-fluorobenzyl 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-({N-[({(2S)-1-[(3-fluorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-fluorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9B0M
 
 | |
8UR1
 
 | |
9ASY
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-chlorobenzyl 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9AT7
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a 2,2-difluoro-5-methylbenzo[1,3]dioxole 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9AT6
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylbicyclo[2.2.1]heptene 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9ASZ
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenylethyl 2-pyrrolidone inhibitor | | 分子名称: | (1S,2S)-1-hydroxy-2-{[N-({[(2S)-5-oxo-1-(2-phenylethyl)pyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
8VW0
 
 | |
9AT5
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a 1-methyl-4,4-difluorocyclohexyl 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9AT1
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (R-enantiomer) | | 分子名称: | (1S,2S)-2-{[N-({[(2R)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, TETRAETHYLENE GLYCOL | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9AT4
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylbicyclo[2.2.1]heptane 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-{[N-({[(2S)-1-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9AT0
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (S-enantiomer) | | 分子名称: | (1R,2S)-2-{[N-({[(2S)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
8VW2
 
 | |
8VW1
 
 | |
7LQV
 
 | | Cryo-EM structure of NTD-directed neutralizing antibody 4-8 Fab in complex with SARS-CoV-2 S2P spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-8 Heavy Chain, 4-8 Light chain, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2021-02-15 | | 公開日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite
Cell Host Microbe, 2021
|
|
7LQW
 
 | | Cryo-EM structure of NTD-directed neutralizing antibody 2-17 Fab in complex with SARS-CoV-2 S2P spike | | 分子名称: | 2-17 Heavy Chain, 2-17 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2021-02-15 | | 公開日 | 2021-03-24 | | 最終更新日 | 2021-05-26 | | 実験手法 | ELECTRON MICROSCOPY (4.47 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
4LCW
 
 | | The structure of human MAIT TCR in complex with MR1-K43A-RL-6-Me-7OH | | 分子名称: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Patel, O, Rossjohn, J. | | 登録日 | 2013-06-24 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells.
J.Exp.Med., 210, 2013
|
|
