5GUJ
 
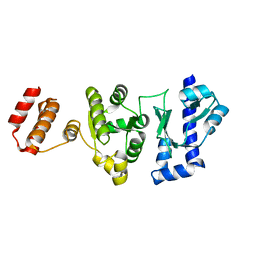 | |
6IQL
 
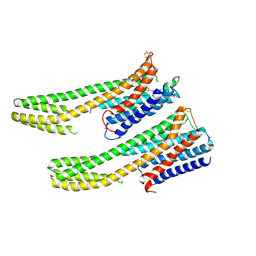 | | Crystal structure of dopamine receptor D4 bound to the subtype-selective ligand, L745870 | | 分子名称: | 3-{[4-(4-chlorophenyl)piperazin-1-yl]methyl}-1H-pyrrolo[2,3-b]pyridine, D(4) dopamine receptor,Soluble cytochrome b562,D(4) dopamine receptor | | 著者 | Zhou, Y, Cao, C, Zhang, X.C. | | 登録日 | 2018-11-08 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Crystal structure of dopamine receptor D4 bound to the subtype selective ligand, L745870.
Elife, 8, 2019
|
|
6KIM
 
 | |
5XOH
 
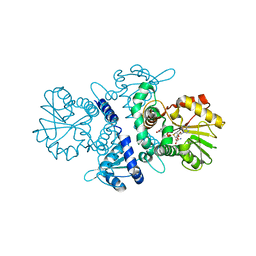 | |
6DCX
 
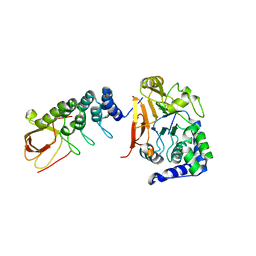 | |
7M42
 
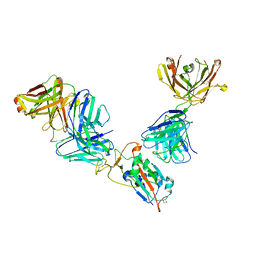 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of neutralizing antibodies REGN10985 and REGN10989 | | 分子名称: | REGN10985 antibody Fab fragment heavy chain, REGN10985 antibody Fab fragment light chain, REGN10989 antibody Fab fragment heavy chain, ... | | 著者 | Zhou, Y, Romero Hernandez, A, Saotome, K, Franklin, M.C. | | 登録日 | 2021-03-19 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies.
Cell, 184, 2021
|
|
6LUI
 
 | | Crystal structure of the SAMD1 WH domain and DNA complex | | 分子名称: | Atherin, DNA (5'-D(*AP*CP*CP*TP*GP*CP*GP*CP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*AP*TP*GP*GP*TP*GP*CP*GP*CP*AP*GP*GP*T)-3') | | 著者 | Zhou, Y, Cao, Y, Wang, Z. | | 登録日 | 2020-01-29 | | 公開日 | 2021-02-03 | | 最終更新日 | 2021-07-07 | | 実験手法 | X-RAY DIFFRACTION (1.781 Å) | | 主引用文献 | The SAM domain-containing protein 1 (SAMD1) acts as a repressive chromatin regulator at unmethylated CpG islands.
Sci Adv, 7, 2021
|
|
3M8L
 
 | |
3RIY
 
 | |
3RIG
 
 | |
3U3D
 
 | |
3U31
 
 | |
6LT9
 
 | |
2HVJ
 
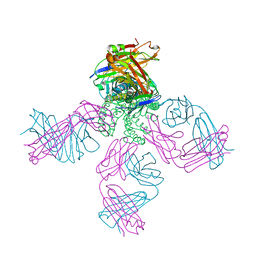 | | Crystal structure of KcsA-Fab-TBA complex in low K+ | | 分子名称: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, NONAN-1-OL, POTASSIUM ION, ... | | 著者 | Zhou, Y. | | 登録日 | 2006-07-28 | | 公開日 | 2007-02-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel.
J.Mol.Biol., 366, 2007
|
|
2HVK
 
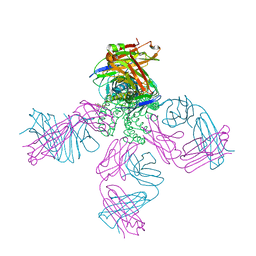 | | crystal structure of the KcsA-Fab-TBA complex in high K+ | | 分子名称: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, Antibody Fab heavy chain, Antibody Fab light chain, ... | | 著者 | Zhou, Y. | | 登録日 | 2006-07-28 | | 公開日 | 2007-02-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel.
J.Mol.Biol., 366, 2007
|
|
7VO5
 
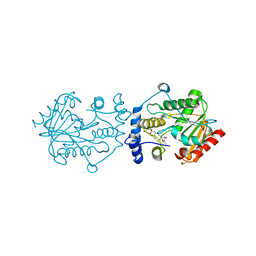 | | Pimaricin type I PKS thioesterase domain (holo Pim TE) | | 分子名称: | (1R,3S,5E,7S,11R,13E,15E,17E,19E,21R,23S,24R,25S)-11,24-dimethyl-1,3,7,21,25-pentakis(oxidanyl)-10,27-dioxabicyclo[21.3.1]heptacosa-5,13,15,17,19-pentaen-9-one, ScnS4 | | 著者 | Bai, L, Zhou, Y. | | 登録日 | 2021-10-12 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and Mechanistic Insights into Chain Release of the Polyene PKS Thioesterase Domain
Acs Catalysis, 12, 2022
|
|
7VO4
 
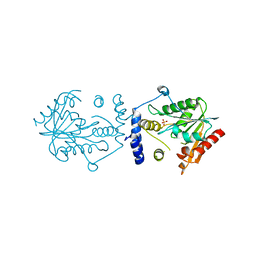 | |
8T2I
 
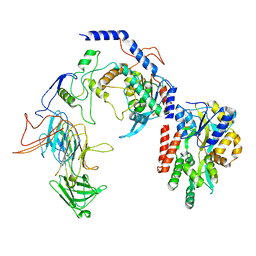 | | Negative stain EM assembly of MYC, JAZ, and NINJA complex | | 分子名称: | AFP homolog 2, Maltose/maltodextrin-binding periplasmic protein, Protein TIFY 10A, ... | | 著者 | Zhou, X.E, Zhang, Y, Zhou, Y, He, Q, Cao, X, Kariapper, L, Suino-Powell, K, Zhu, Y, Zhang, F, Karsten, M. | | 登録日 | 2023-06-06 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (10.4 Å) | | 主引用文献 | Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling.
Plant Commun., 4, 2023
|
|
4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | 分子名称: | Major vault protein | | 著者 | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | 登録日 | 2008-10-24 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
5KLV
 
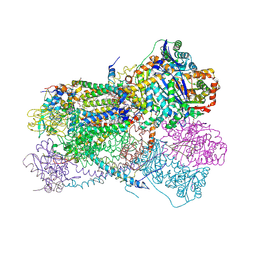 | | Structure of bos taurus cytochrome bc1 with fenamidone inhibited | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | 著者 | Xia, D, Esser, L, Zhou, F, Zhou, Y, Xiao, Y, Tang, W.K, Yu, C.A, Qin, Z. | | 登録日 | 2016-06-25 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.652 Å) | | 主引用文献 | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
2GCQ
 
 | |
8DTI
 
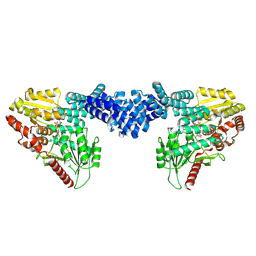 | | Cryo-EM structure of Arabidopsis SPY in complex with GDP-fucose | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | 登録日 | 2022-07-25 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTG
 
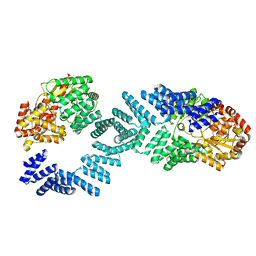 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 1 | | 分子名称: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | 登録日 | 2022-07-25 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTH
 
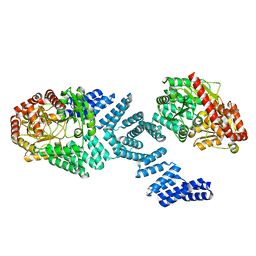 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 2 | | 分子名称: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | 登録日 | 2022-07-25 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTF
 
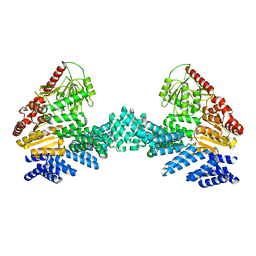 | | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs | | 分子名称: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | 登録日 | 2022-07-25 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
