4Z30
 
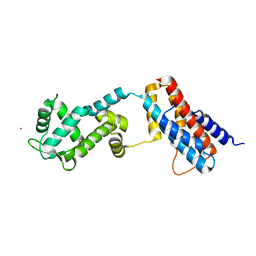 | | Crystal structure of the ROQ domain of human Roquin-2 | | 分子名称: | Roquin-2, UNKNOWN ATOM OR ION | | 著者 | DONG, A, ZHANG, Q, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | 登録日 | 2015-03-30 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | New Insights into the RNA-Binding and E3 Ubiquitin Ligase Activities of Roquins.
Sci Rep, 5, 2015
|
|
3O03
 
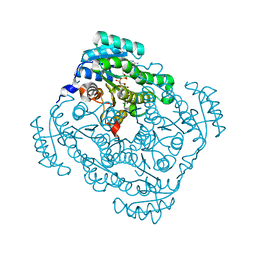 | | Quaternary complex structure of gluconate 5-dehydrogenase from streptococcus suis type 2 | | 分子名称: | CALCIUM ION, D-gluconic acid, Dehydrogenase with different specificities, ... | | 著者 | Peng, H, Gao, F, Zhang, Q, Liu, Y, Gao, G.F. | | 登録日 | 2010-07-18 | | 公開日 | 2010-12-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Insight Into the Catalytic Mechanism of Gluconate 5-Dehydrogenase from Streptococcus Suis: Crystal Structures of the Substrate-Free and Quaternary Complex Enzymes.
Protein Sci., 18, 2009
|
|
4QN1
 
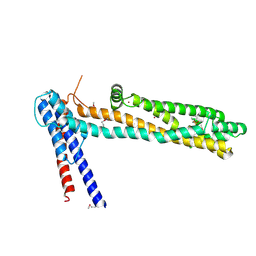 | | Crystal Structure of a Functionally Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH | | 分子名称: | E3 ubiquitin-protein ligase SHPRH, SULFATE ION, UNKNOWN ATOM OR ION, ... | | 著者 | Dong, A, Zhang, Q, Li, Y, Walker, J.R, Guan, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | 登録日 | 2014-06-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Crystal structure of a Function Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH
To be Published
|
|
3CXR
 
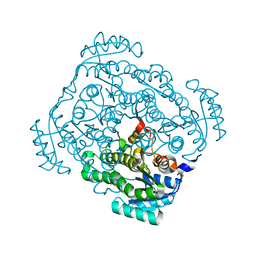 | | Crystal structure of gluconate 5-dehydrogase from streptococcus suis type 2 | | 分子名称: | Dehydrogenase with different specificities | | 著者 | Peng, H, Gao, F, Zhang, Q, Liu, Y, Gao, G.F. | | 登録日 | 2008-04-25 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insight into the catalytic mechanism of gluconate 5-dehydrogenase from Streptococcus suis: Crystal structures of the substrate-free and quaternary complex enzymes.
Protein Sci., 18, 2009
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | 分子名称: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | 著者 | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | 登録日 | 2011-12-13 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | 分子名称: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | 著者 | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | 登録日 | 2011-12-14 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
7CHO
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-1D2 with RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-1D2 heavy chain, ... | | 著者 | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | 登録日 | 2020-07-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.561 Å) | | 主引用文献 | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHS
 
 | | Crystal structure of SARS-CoV-2 antibody P22A-1D1 with RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P22A-1D1 heavy chain, ... | | 著者 | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | 登録日 | 2020-07-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHP
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-3C8 with RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-3C8 heavy chain, ... | | 著者 | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | 登録日 | 2020-07-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.357 Å) | | 主引用文献 | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
5H9M
 
 | | Crystal structure of siah2 SBD domain | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase SIAH2, PENTAETHYLENE GLYCOL, ... | | 著者 | Dong, A, Zhang, Q, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | 登録日 | 2015-12-28 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.761 Å) | | 主引用文献 | Crystal structure of siah2 SBD domain
to be published
|
|
6IIC
 
 | | CryoEM structure of Mud Crab Dicistrovirus | | 分子名称: | VP1 of Mud crab dicistrovirus, VP2 of Mud crab dicistrovirus, VP3 of Mud crab dicistrovirus, ... | | 著者 | Zhang, Q, Gao, Y. | | 登録日 | 2018-10-04 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-electron Microscopy Structures of Novel Viruses from Mud CrabScylla paramamosainwith Multiple Infections.
J. Virol., 93, 2019
|
|
6IZL
 
 | |
6LI6
 
 | |
6JDD
 
 | | Crystal structure of the cypemycin decarboxylase CypD. | | 分子名称: | Cypemycin cysteine dehydrogenase (decarboxylating), DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Zhang, Q, Yuan, H. | | 登録日 | 2019-02-01 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Convergent evolution of the Cys decarboxylases involved in aminovinyl-cysteine (AviCys) biosynthesis.
FEBS Lett., 593, 2019
|
|
7VOI
 
 | | Structure of the human CNOT1(MIF4G)-CNOT6L-CNOT7 complex | | 分子名称: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 6-like, CCR4-NOT transcription complex subunit 7 | | 著者 | Bartlam, M, Zhang, Q. | | 登録日 | 2021-10-13 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (4.38 Å) | | 主引用文献 | Structure of the human Ccr4-Not nuclease module using X-ray crystallography and electron paramagnetic resonance spectroscopy distance measurements.
Protein Sci., 31, 2022
|
|
3MFQ
 
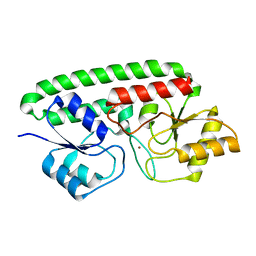 | | A Glance into the Metal Binding Specificity of TroA: Where Elaborate Behaviors Occur in the Active Center | | 分子名称: | High-affinity zinc uptake system protein znuA, ZINC ION | | 著者 | Gao, G.F, Zheng, B, Zhang, Q, Gao, J, Han, H, Li, M. | | 登録日 | 2010-04-03 | | 公開日 | 2011-04-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Insight into the interaction of metal ions with TroA from Streptococcus suis
Plos One, 6, 2011
|
|
2RNW
 
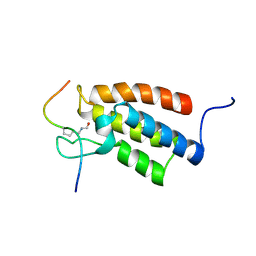 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the Human Transcriptional Co-Activators PCAf and CBP | | 分子名称: | Histone H3, Histone acetyltransferase PCAF | | 著者 | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | 登録日 | 2008-02-03 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNX
 
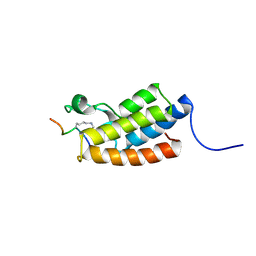 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | 分子名称: | Histone H3, Histone acetyltransferase PCAF | | 著者 | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | 登録日 | 2008-02-03 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
3MKT
 
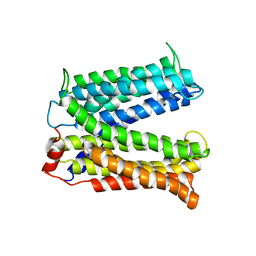 | | Structure of a Cation-bound Multidrug and Toxin Compound Extrusion (MATE) transporter | | 分子名称: | Multi antimicrobial extrusion protein (Na(+)/drug antiporter) MATE-like MDR efflux pump | | 著者 | He, X, Szewczyk, P, Karyakin, A, Evin, M, Hong, W.-X, Zhang, Q, Chang, G. | | 登録日 | 2010-04-15 | | 公開日 | 2010-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Structure of a cation-bound multidrug and toxic compound extrusion transporter.
Nature, 467, 2010
|
|
3MKU
 
 | | Structure of a Cation-bound Multidrug and Toxin Compound Extrusion (MATE) transporter | | 分子名称: | Multi antimicrobial extrusion protein (Na(+)/drug antiporter) MATE-like MDR efflux pump, RUBIDIUM ION | | 著者 | He, X, Szewczyk, P, Karyakin, A, Evin, M, Hong, W.-X, Zhang, Q, Chang, G. | | 登録日 | 2010-04-15 | | 公開日 | 2010-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Structure of a cation-bound multidrug and toxic compound extrusion transporter.
Nature, 467, 2010
|
|
1F3C
 
 | |
1F95
 
 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND BIM PEPTIDE COMPLEX | | 分子名称: | BCL2-LIKE 11 (APOPTOSIS FACILITATOR), DYNEIN | | 著者 | Fan, J.-S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | 登録日 | 2000-07-07 | | 公開日 | 2001-02-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
1F96
 
 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND NNOS PEPTIDE COMPLEX | | 分子名称: | DYNEIN LIGHT CHAIN 8, PROTEIN (NNOS, NEURONAL NITRIC OXIDE SYNTHASE) | | 著者 | Fan, J.S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | 登録日 | 2000-07-07 | | 公開日 | 2001-02-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
6DNY
 
 | |
4V8M
 
 | | High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome | | 分子名称: | 18S RRNA OF THE SMALL RIBOSOMAL SUBUNIT, 40S RIBOSOMAL PROTEIN S10, PUTATIVE, ... | | 著者 | Hashem, Y, des Georges, A, Fu, J, Buss, S.N, Jossinet, F, Jobe, A, Zhang, Q, Liao, H.Y, Grassucci, R.A, Bajaj, C, Westhof, E, Madison-Antenucci, S, Frank, J. | | 登録日 | 2012-12-09 | | 公開日 | 2014-07-09 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (5.57 Å) | | 主引用文献 | High-Resolution Cryo-Electron Microscopy Structure of the Trypanosoma Brucei Ribosome.
Nature, 494, 2013
|
|
