8TJI
 
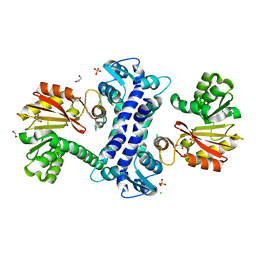 | | SAM-dependent methyltransferase RedM, apo | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, RedM, ... | | 著者 | Daniel-Ivad, P, Ryan, K.S. | | 登録日 | 2023-07-22 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
8TJJ
 
 | |
8TJK
 
 | |
4IXM
 
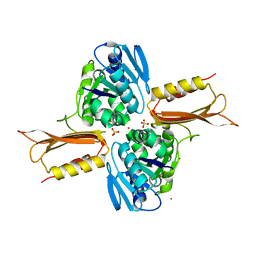 | | Crystal structure of Zn(II)-bound YjiA GTPase from E. coli | | 分子名称: | SULFATE ION, Uncharacterized GTP-binding protein YjiA, ZINC ION | | 著者 | Jost, M, Ryan, K.S, Turo, K.E, Drennan, C.L. | | 登録日 | 2013-01-26 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases.
Biochemistry, 52, 2013
|
|
8U06
 
 | |
8U05
 
 | |
8U07
 
 | |
7LR2
 
 | |
7LR7
 
 | |
7LR6
 
 | |
7LQX
 
 | |
7LR1
 
 | |
7LR8
 
 | |
7JQ2
 
 | |
7JPY
 
 | |
7JQ4
 
 | |
7JQ0
 
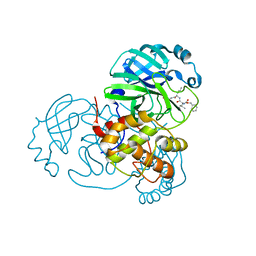 | |
7JPZ
 
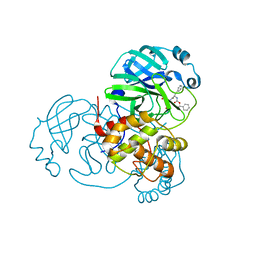 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI1 | | 分子名称: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase | | 著者 | Yang, K, Liu, W. | | 登録日 | 2020-08-10 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ5
 
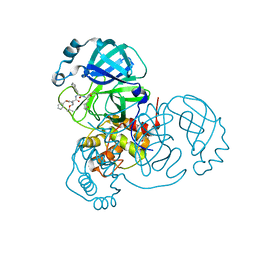 | |
7JQ3
 
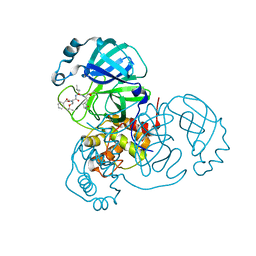 | |
7JQ1
 
 | |
5UN5
 
 | |
5UN6
 
 | |
4ZBL
 
 | |
7M2P
 
 | |
