1GE6
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | 分子名称: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | 著者 | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | 登録日 | 2000-10-11 | | 公開日 | 2001-03-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GE5
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | 分子名称: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | 著者 | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | 登録日 | 2000-10-11 | | 公開日 | 2001-03-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G12
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | 分子名称: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | 著者 | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | 登録日 | 2000-10-10 | | 公開日 | 2001-03-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GE7
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | 分子名称: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | 著者 | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | 登録日 | 2000-10-11 | | 公開日 | 2001-03-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6JTB
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with citrate from Porphyromonas gingivalis (Space) | | 分子名称: | Asp/Glu-specific dipeptidyl-peptidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | 登録日 | 2019-04-10 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
6JTC
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with SH-5 from Porphyromonas gingivalis (Space) | | 分子名称: | 2-(2-azanylethylamino)-5-nitro-benzoic acid, Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | 登録日 | 2019-04-10 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
5H41
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose, isofagomine, sulfate ion | | 分子名称: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, SULFATE ION, Uncharacterized protein, ... | | 著者 | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | 登録日 | 2016-10-28 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5H40
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose | | 分子名称: | CALCIUM ION, GLYCEROL, Uncharacterized protein, ... | | 著者 | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | 登録日 | 2016-10-28 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5H3Z
 
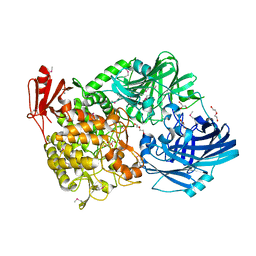 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | 登録日 | 2016-10-28 | | 公開日 | 2017-03-01 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5H42
 
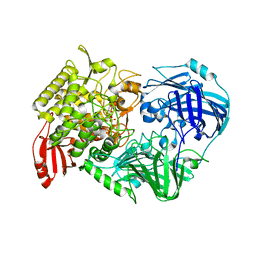 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with alpha-d-glucose-1-phosphate | | 分子名称: | 1-O-phosphono-alpha-D-glucopyranose, Uncharacterized protein, alpha-D-glucopyranose | | 著者 | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | 登録日 | 2016-10-28 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
1CCR
 
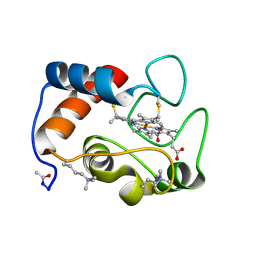 | | STRUCTURE OF RICE FERRICYTOCHROME C AT 2.0 ANGSTROMS RESOLUTION | | 分子名称: | CYTOCHROME C, HEME C | | 著者 | Ochi, H, Hata, Y, Tanaka, N, Kakudo, M, Sakurai, T, Aihara, S, Morita, Y. | | 登録日 | 1983-03-14 | | 公開日 | 1983-04-21 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of rice ferricytochrome c at 2.0 A resolution.
J.Mol.Biol., 166, 1983
|
|
1G2U
 
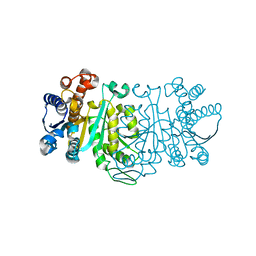 | | THE STRUCTURE OF THE MUTANT, A172V, OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THERMUS THERMOPHILUS HB8 : ITS THERMOSTABILITY AND STRUCTURE. | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | 登録日 | 2000-10-21 | | 公開日 | 2000-11-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GC8
 
 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO PHE | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | 登録日 | 2000-07-27 | | 公開日 | 2000-09-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GC9
 
 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO GLY | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | 登録日 | 2000-07-28 | | 公開日 | 2000-09-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DR0
 
 | | STRUCTURE OF MODIFIED 3-ISOPROPYLMALATE DEHYDROGENASE AT THE C-TERMINUS, HD708 | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Nurachman, Z, Akanuma, S, Sato, T, Oshima, T, Tanaka, N. | | 登録日 | 2000-01-06 | | 公開日 | 2000-01-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of 3-isopropylmalate dehydrogenases with mutations at the C-terminus: crystallographic analyses of structure-stability relationships.
Protein Eng., 13, 2000
|
|
1DR8
 
 | | STRUCTURE OF MODIFIED 3-ISOPROPYLMALATE DEHYDROGENASE AT THE C-TERMINUS, HD177 | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Nurachman, Z, Akanuma, S, Sato, T, Oshima, T, Tanaka, N. | | 登録日 | 2000-01-06 | | 公開日 | 2000-01-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of 3-isopropylmalate dehydrogenases with mutations at the C-terminus: crystallographic analyses of structure-stability relationships.
Protein Eng., 13, 2000
|
|
1DPZ
 
 | | STRUCTURE OF MODIFIED 3-ISOPROPYLMALATE DEHYDROGENASE AT THE C-TERMINUS, HD711 | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Nurachman, Z, Akanuma, S, Sato, T, Oshima, T, Tanaka, N. | | 登録日 | 1999-12-29 | | 公開日 | 2000-01-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of 3-isopropylmalate dehydrogenases with mutations at the C-terminus: crystallographic analyses of structure-stability relationships.
Protein Eng., 13, 2000
|
|
8JNW
 
 | | PfDXR - Mn2+ - NADPH - MAMK89 quaternary complex | | 分子名称: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | 著者 | Takada, S, Sakamoto, Y, Tanaka, N. | | 登録日 | 2023-06-08 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Reverse N -Substituted Hydroxamic Acid Derivatives of Fosmidomycin Target a Previously Unknown Subpocket of 1-Deoxy-d-xylulose 5-Phosphate Reductoisomerase (DXR).
Acs Infect Dis., 10, 2024
|
|
8JNV
 
 | | PfDXR - Mn2+ - MAMK89 ternary complex | | 分子名称: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | 著者 | Takada, S, Sakamoto, Y, Tanaka, N. | | 登録日 | 2023-06-06 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Reverse N -Substituted Hydroxamic Acid Derivatives of Fosmidomycin Target a Previously Unknown Subpocket of 1-Deoxy-d-xylulose 5-Phosphate Reductoisomerase (DXR).
Acs Infect Dis., 10, 2024
|
|
5AXD
 
 | |
5AXC
 
 | | Crystal structure of mouse SAHH complexed with 3'-keto aristeromycin | | 分子名称: | (2S,3R,5R)-3-(6-amino-9H-purin-9-yl)-2-hydroxy-5-(hydroxymethyl)cyclopentanone, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Kusakabe, Y, Ishihara, M, Tanaka, N. | | 登録日 | 2015-07-24 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of mouse SAHH complexed with 3'-keto aristeromycin
To Be Published
|
|
5AXA
 
 | |
5AXB
 
 | | Crystal structure of mouse SAHH complexed with noraristeromycin | | 分子名称: | (1S,2R,3S,4R)-4-(6-aminopurin-9-yl)cyclopentane-1,2,3-triol, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Kusakabe, Y, Ishihara, M, Tanaka, N. | | 登録日 | 2015-07-24 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of mouse SAHH complexed with noraristeromycin
To Be Published
|
|
3VP8
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p | | 分子名称: | General transcriptional corepressor TUP1 | | 著者 | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | 登録日 | 2012-02-28 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
1IPD
 
 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | 著者 | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | 登録日 | 1992-01-29 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
