2Z1Y
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (complexed with N-(5'-phosphopyridoxyl)-L-leucine), from Thermus thermophilus HB27 | | 分子名称: | Alpha-aminodipate aminotransferase, LEUCINE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2007-05-16 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Mechanism of broad substrate specificity of alpha-aminoadipate aminotransferase from Thermus thermophilus
To be Published
|
|
2ZP7
 
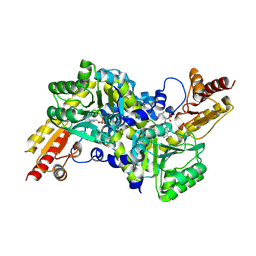 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (Leucine complex), from Thermus thermophilus HB27 | | 分子名称: | Alpha-aminodipate aminotransferase, LEUCINE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2008-06-30 | | 公開日 | 2009-01-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Mechanism for multiple-substrates recognition of alpha-aminoadipate aminotransferase from Thermus thermophilus
Proteins, 2008
|
|
3CBF
 
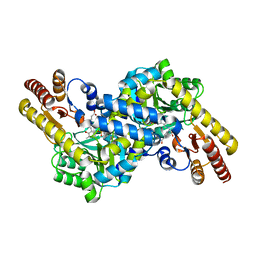 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase, from Thermus thermophilus HB27 | | 分子名称: | (2S)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]hexanedioic acid, Alpha-aminodipate aminotransferase | | 著者 | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2008-02-21 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Mechanism for multiple-substrates recognition of alpha-aminoadipate aminotransferase from Thermus thermophilus
Proteins, 2008
|
|
7WLG
 
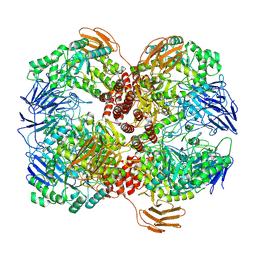 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | 分子名称: | Alpha-xylosidase | | 著者 | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | 登録日 | 2022-01-13 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJE
 
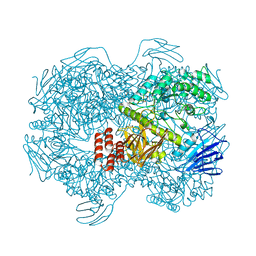 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerotetraose | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJD
 
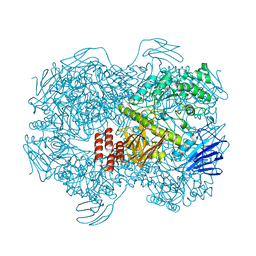 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerotriose | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, ... | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJF
 
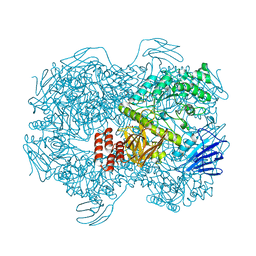 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with kojibiose | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-2)-alpha-D-glucopyranose | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJB
 
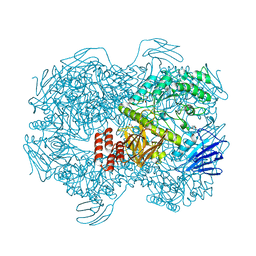 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase in complex with glucose | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose, ... | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJC
 
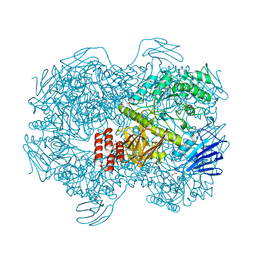 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerose | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJ9
 
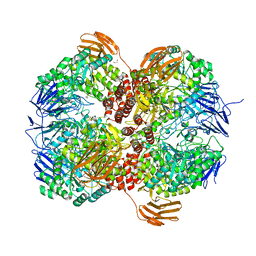 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase, P21 space group | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, Xylitol | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJA
 
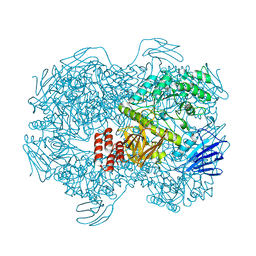 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase, P6322 space group | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7DAA
 
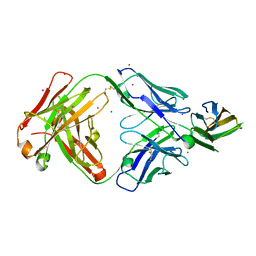 | | Crystal structure of basigin complexed with anti-basigin Fab fragment | | 分子名称: | CADMIUM ION, Heavy chain of antibody Fab fragment, Isoform 2 of Basigin, ... | | 著者 | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | 登録日 | 2020-10-16 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D9Z
 
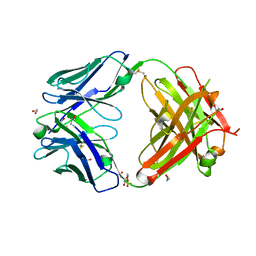 | | Crystal structure of anti-basigin Fab fragment | | 分子名称: | 1,2-ETHANEDIOL, CITRATE ANION, Heavy chain of antibody Fab fragment, ... | | 著者 | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | 登録日 | 2020-10-14 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.123 Å) | | 主引用文献 | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7DCE
 
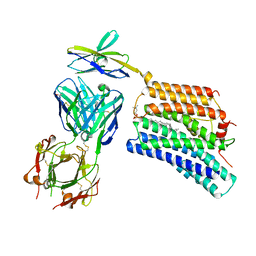 | | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | | 分子名称: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Heavy chain of Fab fragment, Isoform 2 of Basigin, ... | | 著者 | Sakuragi, T, Kanai, R, Tsutsumi, A, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Kikkawa, M, Toyoshima, C, Nagata, S. | | 登録日 | 2020-10-26 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7FE3
 
 | |
7FE4
 
 | |
3A64
 
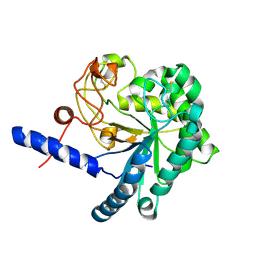 | | Crystal structure of CcCel6C, a glycoside hydrolase family 6 enzyme, from Coprinopsis cinerea | | 分子名称: | Cellobiohydrolase, MAGNESIUM ION | | 著者 | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | 登録日 | 2009-08-21 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3ABX
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with p-nitrophenyl beta-D-cellotrioside | | 分子名称: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | 著者 | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | 登録日 | 2009-12-24 | | 公開日 | 2010-01-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3A9B
 
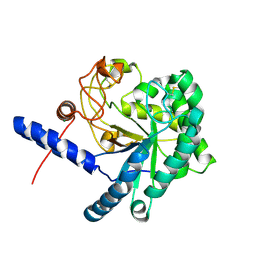 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with cellobiose | | 分子名称: | Cellobiohydrolase, MAGNESIUM ION, beta-D-glucopyranose, ... | | 著者 | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | 登録日 | 2009-10-22 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3VSS
 
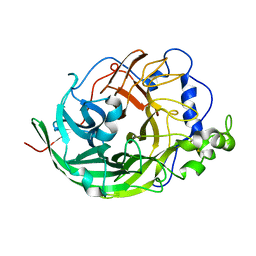 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain complexed with fructose | | 分子名称: | Beta-fructofuranosidase, beta-D-fructofuranose | | 著者 | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | 登録日 | 2012-05-08 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3VSR
 
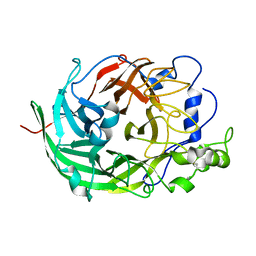 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain | | 分子名称: | Beta-fructofuranosidase | | 著者 | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | 登録日 | 2012-05-08 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3WJL
 
 | | Crystal structure of IIb selective Fc variant, Fc(V12), in complex with FcgRIIb | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, Low affinity immunoglobulin gamma Fc region receptor II-c, ... | | 著者 | Kadono, S, Mimoto, F, Katada, H, Igawa, T, Kuramochi, T, Muraoka, M, Wada, Y, Haraya, K, Miyazaki, T, Hattori, K. | | 登録日 | 2013-10-11 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Engineered antibody Fc variant with selectively enhanced Fc gamma RIIb binding over both Fc gamma RIIaR131 and Fc gamma RIIaH131.
Protein Eng.Des.Sel., 26, 2013
|
|
3WJJ
 
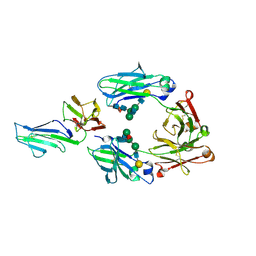 | | Crystal structure of IIb selective Fc variant, Fc(P238D), in complex with FcgRIIb | | 分子名称: | Ig gamma-1 chain C region, Low affinity immunoglobulin gamma Fc region receptor II-b, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Kadono, S, Mimoto, F, Katada, H, Igawa, T, Kuramochi, T, Muraoka, M, Wada, Y, Haraya, K, Miyazaki, T, Hattori, K. | | 登録日 | 2013-10-10 | | 公開日 | 2013-11-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Engineered antibody Fc variant with selectively enhanced Fc gamma RIIb binding over both Fc gamma RIIaR131 and Fc gamma RIIaH131.
Protein Eng.Des.Sel., 26, 2013
|
|
3WPU
 
 | | Full-length beta-fructofuranosidase from Microbacterium saccharophilum K-1 | | 分子名称: | Beta-fructofuranosidase, GLYCEROL | | 著者 | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | 登録日 | 2014-01-17 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WPV
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/F447V/F470Y/P500S | | 分子名称: | Beta-fructofuranosidase, GLYCEROL | | 著者 | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | 登録日 | 2014-01-17 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
