5MW1
 
 | |
5LY3
 
 | |
5LJN
 
 | | Structure of the HOIP PUB domain bound to SPATA2 PIM peptide | | 分子名称: | E3 ubiquitin-protein ligase RNF31, GLYCEROL, SULFATE ION, ... | | 著者 | Elliott, P.R, Komander, D. | | 登録日 | 2016-07-18 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
6UKA
 
 | | Crystal structure of RHOG and ELMO complex | | 分子名称: | Engulfment and cell motility protein 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Jo, C.H, Killoran, R.C, Smith, M.J. | | 登録日 | 2019-10-04 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
6QAJ
 
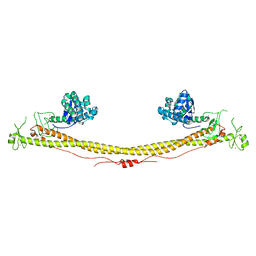 | | Structure of the tripartite motif of KAP1/TRIM28 | | 分子名称: | Endolysin,Transcription intermediary factor 1-beta, ZINC ION | | 著者 | Stoll, G.A, Oda, S, Yu, M, Modis, Y. | | 登録日 | 2018-12-19 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.901 Å) | | 主引用文献 | Structure of KAP1 tripartite motif identifies molecular interfaces required for retroelement silencing.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7Q6D
 
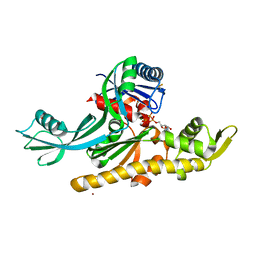 | | E. coli FtsA 1-405 ATP 3 Ni | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION, ... | | 著者 | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | 登録日 | 2021-11-06 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7Q6I
 
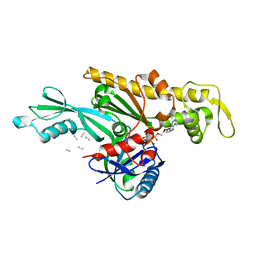 | | Vibrio maritimus FtsA 1-396 ATP and FtsN 1-29, bent tetramers in double filament arrangement | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, Cell division protein FtsN (polyAla model), ... | | 著者 | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | 登録日 | 2021-11-07 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7Q6G
 
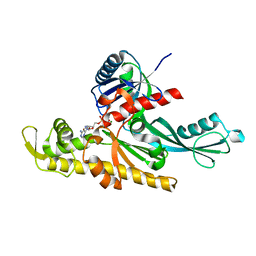 | |
7Q6F
 
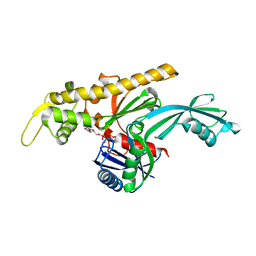 | | Vibrio maritimus FtsA 1-396 ATP, double filament | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | 著者 | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | 登録日 | 2021-11-07 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
6RIB
 
 | |
6RIA
 
 | |
6S53
 
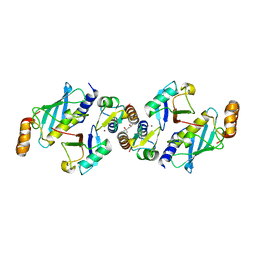 | | Crystal structure of TRIM21 RING domain in complex with an isopeptide-linked Ube2N~ubiquitin conjugate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, ... | | 著者 | Kiss, L, Boland, A, Neuhaus, D, James, L.C. | | 登録日 | 2019-06-30 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases.
Nat Commun, 10, 2019
|
|
4CKP
 
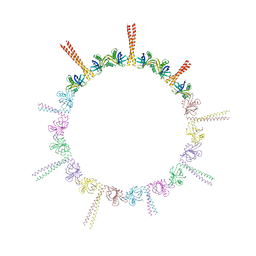 | |
4CKM
 
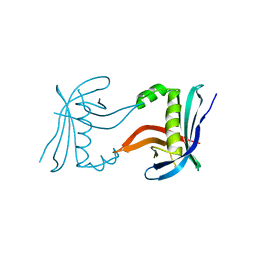 | |
4CKN
 
 | |
4P80
 
 | | Structure of ancestral PyrR protein (AncGREENPyrR) | | 分子名称: | Ancestral PyrR protein (Green), SULFATE ION | | 著者 | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | 登録日 | 2014-03-29 | | 公開日 | 2014-12-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P83
 
 | | Structure of engineered PyrR protein (PURPLE PyrR) | | 分子名称: | Engineered PyrR protein (Purple), URIDINE-5'-MONOPHOSPHATE | | 著者 | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | 登録日 | 2014-03-30 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P86
 
 | | Structure of PyrR protein from Bacillus subtilis with GMP | | 分子名称: | Bifunctional protein PyrR, GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE | | 著者 | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | 登録日 | 2014-03-30 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P84
 
 | | Structure of engineered PyrR protein (VIOLET PyrR) | | 分子名称: | Bifunctional protein PyrR, GLYCEROL, SULFATE ION | | 著者 | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | 登録日 | 2014-03-30 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P81
 
 | | Structure of ancestral PyrR protein (AncORANGEPyrR) | | 分子名称: | Ancestral PyrR protein (Orange), GLYCEROL, SULFATE ION | | 著者 | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | 登録日 | 2014-03-29 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P82
 
 | | Structure of PyrR protein from Bacillus subtilis | | 分子名称: | Bifunctional protein PyrR, SULFATE ION | | 著者 | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | 登録日 | 2014-03-30 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P3K
 
 | | Structure of ancestral PyrR protein (PLUMPyrR) | | 分子名称: | Ancestral PyrR protein (Plum), PENTAETHYLENE GLYCOL, SODIUM ION, ... | | 著者 | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | 登録日 | 2014-03-08 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
8A2Q
 
 | |
6H9O
 
 | |
6H9N
 
 | |
