7CKA
 
 | |
7EWH
 
 | | Crystal structure of human PHGDH in complex with Homoharringtonine | | 分子名称: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, D-3-phosphoglycerate dehydrogenase | | 著者 | Hsieh, C.H, Cheng, Y.S, Lee, Y.S, Huang, H.C, Juan, H.F. | | 登録日 | 2021-05-25 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Homoharringtonine as a PHGDH inhibitor: Unraveling metabolic dependencies and developing a potent therapeutic strategy for high-risk neuroblastoma.
Biomed Pharmacother, 166, 2023
|
|
2DW5
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with N-alpha-benzoyl-N5-(2-fluoro-1-iminoethyl)-L-ornithine amide | | 分子名称: | CALCIUM ION, N-[(1S)-1-(AMINOCARBONYL)-4-(ETHANIMIDOYLAMINO)BUTYL]BENZAMIDE, Protein-arginine deiminase type-4, ... | | 著者 | Luo, Y, Arita, K, Sato, M, Thompson, P.R. | | 登録日 | 2006-08-04 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Inhibitors and Inactivators of Protein Arginine Deiminase 4: Functional and Structural Characterization
Biochemistry, 45, 2006
|
|
2BIF
 
 | | 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE H256A MUTANT WITH F6P IN PHOSPHATASE ACTIVE SITE | | 分子名称: | 6-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Yuen, M.H, Hasemann, C.A. | | 登録日 | 1998-10-26 | | 公開日 | 1999-02-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the H256A mutant of rat testis fructose-6-phosphate,2-kinase/fructose-2,6-bisphosphatase. Fructose 6-phosphate in the active site leads to mechanisms for both mutant and wild type bisphosphatase activities.
J.Biol.Chem., 274, 1999
|
|
4MA4
 
 | | S-glutathionylated PFKFB3 | | 分子名称: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ACETIC ACID, ... | | 著者 | Lee, Y.-H, Seo, M.-S. | | 登録日 | 2013-08-15 | | 公開日 | 2013-12-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | PFKFB3 Regulates Oxidative Stress Homeostasis via Its S-Glutathionylation in Cancer.
J.Mol.Biol., 426, 2014
|
|
3ZXO
 
 | | CRYSTAL STRUCTURE OF THE MUTANT ATP-BINDING DOMAIN OF MYCOBACTERIUM TUBERCULOSIS DOSS | | 分子名称: | ACETATE ION, GLYCEROL, REDOX SENSOR HISTIDINE KINASE RESPONSE REGULATOR DEVS, ... | | 著者 | Cho, H.Y, Cho, H.J, Kang, B.S. | | 登録日 | 2011-08-13 | | 公開日 | 2011-08-24 | | 最終更新日 | 2013-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Activation of ATP Binding for the Autophosphorylation of Doss, a Mycobacterium Tuberculosis Histidine Kinase Lacking an ATP-Lid Motif.
J.Biol.Chem., 288, 2013
|
|
3ZXQ
 
 | |
7EPP
 
 | |
4OOU
 
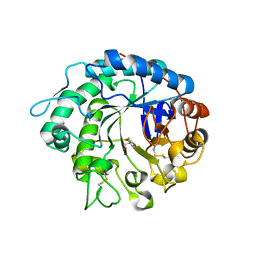 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase | | 著者 | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | 登録日 | 2014-02-04 | | 公開日 | 2014-08-06 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
6J6T
 
 | | Crystal Structure of HDA15 HD domain | | 分子名称: | Histone deacetylase 15, POTASSIUM ION, SULFATE ION, ... | | 著者 | Cheng, Y.S, Hsu, J.C, Hung, H.C, Liu, T.C. | | 登録日 | 2019-01-15 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structure of Arabidopsis HISTONE DEACETYLASE15.
Plant Physiol., 184, 2020
|
|
5H4U
 
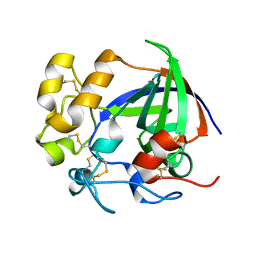 | | Crystal structure of cellulase from Antarctic springtail, Cryptopygus antarcticus | | 分子名称: | Endo-beta-1,4-glucanase | | 著者 | An, Y.J, Hong, S.K, Cha, S.S. | | 登録日 | 2016-11-02 | | 公開日 | 2017-03-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Genetic and Structural Characterization of a Thermo-Tolerant, Cold-Active, and Acidic Endo-beta-1,4-glucanase from Antarctic Springtail, Cryptopygus antarcticus.
J. Agric. Food Chem., 65, 2017
|
|
5AUI
 
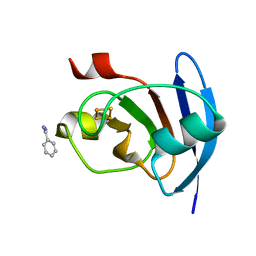 | |
5AUK
 
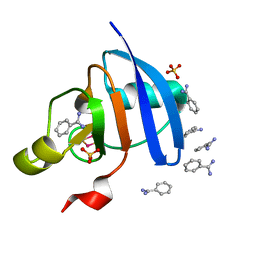 | | Crystal structure of the Ga-substituted Ferredoxin | | 分子名称: | BENZAMIDINE, Ferredoxin-1, SULFATE ION, ... | | 著者 | Kurisu, G, Muraki, N, Taya, M, Hase, T. | | 登録日 | 2015-04-24 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | X-ray Structure and Nuclear Magnetic Resonance Analysis of the Interaction Sites of the Ga-Substituted Cyanobacterial Ferredoxin
Biochemistry, 54, 2015
|
|
5GIW
 
 | | Solution NMR structure of Humanin containing a D-isomerized serine residue | | 分子名称: | Humanin | | 著者 | Furuita, K, Sugiki, T, Alsanousi, N, Fujiwara, T, Kojima, C. | | 登録日 | 2016-06-25 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure and inhibitory effect against amyloid-beta fibrillation of Humanin containing a d-isomerized serine residue
Biochem.Biophys.Res.Commun., 477, 2016
|
|
7FIX
 
 | |
7W5C
 
 | | Crystal structure of Mitogen Activated Protein Kinase 4 (MPK4) from Arabidopsis thaliana | | 分子名称: | MAGNESIUM ION, Mitogen-activated protein kinase 4, Mitogen-activated protein kinase kinase 1, ... | | 著者 | Arold, S.T, Hameed, U.F.S. | | 登録日 | 2021-11-30 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Essential role of the CD docking motif of MPK4 in plant immunity, growth, and development.
New Phytol., 239, 2023
|
|
5XYO
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122G mutant | | 分子名称: | CHLORIDE ION, Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, ... | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYP
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122R mutant | | 分子名称: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYT
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., H130Y mutant | | 分子名称: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, SULFATE ION | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72. | | 分子名称: | CHLORIDE ION, Endotype 6-aminohexanoat-oligomer hydrolase, GLYCEROL, ... | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-07 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYS
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122V mutant | | 分子名称: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5Y0L
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122G/H130Y mutant | | 分子名称: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, SODIUM ION, ... | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-18 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.385 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5Y0M
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D36A/D122G/H130Y/E263Q mutant | | 分子名称: | CHLORIDE ION, Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, ... | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-18 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYQ
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122K mutant | | 分子名称: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5ZF0
 
 | | X-ray Structure of the Electron Transfer Complex between Ferredoxin and Photosystem I | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Kubota-Kawai, H, Mutoh, R, Shinmura, K, Setif, P, Nowaczyk, M, Roegner, M, Ikegami, T, Tanaka, T, Kurisu, G. | | 登録日 | 2018-03-01 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | X-ray structure of an asymmetrical trimeric ferredoxin-photosystem I complex
Nat Plants, 4, 2018
|
|
