6KK6
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 16 | | 分子名称: | 1-[(5~{R},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-5-methyl-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, GLYCEROL, NS3 protease, ... | | 著者 | Quek, J.P. | | 登録日 | 2019-07-23 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6KK5
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 15 | | 分子名称: | 1-[(5~{S},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-5-methyl-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | 著者 | Quek, J.P. | | 登録日 | 2019-07-23 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6KK4
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 9 | | 分子名称: | 1-[(9~{R},16~{S},19~{S})-16,19-bis(4-azanylbutyl)-4,8,15,18,21-pentakis(oxidanylidene)-3,7,14,17,20-pentazabicyclo[21.3.1]heptacosa-1(26),23(27),24-trien-9-yl]guanidine, GLYCEROL, NS3 protease, ... | | 著者 | Quek, J.P. | | 登録日 | 2019-07-23 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6KK2
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 2 | | 分子名称: | 1-[(10~{S},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.2.2]octacosa-1(27),24(28),25-trien-10-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | 著者 | Quek, J.P. | | 登録日 | 2019-07-23 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6KPQ
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 8 | | 分子名称: | 1-[(10~{R},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.3.1]octacosa-1(27),24(28),25-trien-10-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | 著者 | Quek, J.P. | | 登録日 | 2019-08-15 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6KK3
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 4 | | 分子名称: | 1-[(10~{R},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.2.2]octacosa-1(26),24,27-trien-10-yl]guanidine, Genome polyprotein | | 著者 | Quek, J.P. | | 登録日 | 2019-07-23 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
5NZC
 
 | |
5NZB
 
 | |
6T5L
 
 | |
6T5K
 
 | |
5M15
 
 | |
5M13
 
 | | Synthetic nanobody in complex with MBP | | 分子名称: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein, synthetic Nanobody L2_C06 (a-MBP#2) | | 著者 | Zimmermann, I, Egloff, P, Seeger, M.A. | | 登録日 | 2016-10-07 | | 公開日 | 2017-11-15 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.372 Å) | | 主引用文献 | Synthetic single domain antibodies for the conformational trapping of membrane proteins.
Elife, 7, 2018
|
|
5M14
 
 | |
8BCH
 
 | |
8BC9
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 24 | | 分子名称: | 1,2-ETHANEDIOL, N-hydroxybenzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | 著者 | Vester, K, Loll, B, Wahl, M.C. | | 登録日 | 2022-10-15 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCC
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 39 | | 分子名称: | 1,2-ETHANEDIOL, 3-oxidanylbenzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | 著者 | Vester, K, Loll, B, Wahl, M.C. | | 登録日 | 2022-10-15 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCB
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 34 | | 分子名称: | 1,2-ETHANEDIOL, Pre-mRNA-processing-splicing factor 8, SULFANILAMIDE, ... | | 著者 | Vester, K, Loll, B, Wahl, M.C. | | 登録日 | 2022-10-15 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCA
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 26 | | 分子名称: | 1,2-ETHANEDIOL, 3-azanyl-~{N}-methyl-4-(methylamino)benzenesulfonamide, GLYCEROL, ... | | 著者 | Vester, K, Loll, B, Wahl, M.C. | | 登録日 | 2022-10-15 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCE
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 76 | | 分子名称: | 1,2-ETHANEDIOL, N-methoxybenzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | 著者 | Vester, K, Loll, B, Wahl, M.C. | | 登録日 | 2022-10-15 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCF
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 78 | | 分子名称: | 1,2-ETHANEDIOL, Pre-mRNA-processing-splicing factor 8, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | 著者 | Vester, K, Loll, B, Wahl, M.C. | | 登録日 | 2022-10-15 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCG
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 86 | | 分子名称: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-methoxy-~{N}-methyl-benzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | 著者 | Vester, K, Loll, B, Wahl, M.C. | | 登録日 | 2022-10-15 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BC8
 
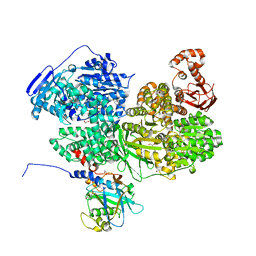 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 18 | | 分子名称: | 1,2-ETHANEDIOL, 3-azanyl-4-oxidanyl-benzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | 著者 | Vester, K, Loll, B, Wahl, M.C. | | 登録日 | 2022-10-15 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCD
 
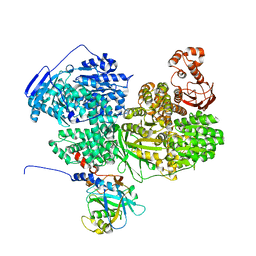 | |
5MCQ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE AND EXOSITE BINDING PEPTIDE INHIBITOR | | 分子名称: | 1,2-ETHANEDIOL, BACE-1 ACTIVE AND EXOSITE BINDING INHIBITOR, Beta-secretase 1 | | 著者 | Kuglstatter, A, Stihle, M, Benz, J. | | 登録日 | 2016-11-10 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MBW
 
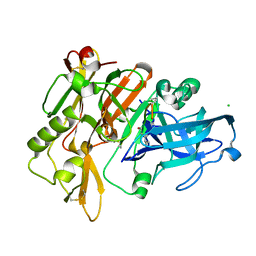 | |
