7DS0
 
 | |
7DRY
 
 | |
7CZ4
 
 | |
1M58
 
 | | Solution Structure of Cytotoxic RC-RNase2 | | 分子名称: | RC-RNase2 ribonuclease | | 著者 | Hsu, C.-H, Liao, Y.-D, Wu, S.-H, Chen, C. | | 登録日 | 2002-07-09 | | 公開日 | 2003-01-09 | | 最終更新日 | 2022-12-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1H, 15N and 13C resonance assignments and secondary structure determination of the RC-RNase 2 from oocytes of bullfrog Rana catesbeiana.
J.Biomol.Nmr, 19, 2001
|
|
2NC5
 
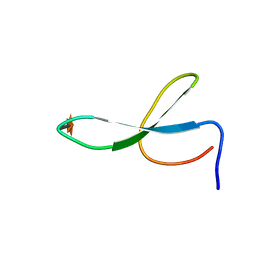 | | Solution Structure of N-Xylosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-D-xylopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC3
 
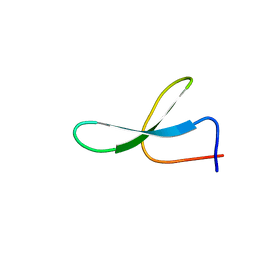 | | Solution Structure of N-Allosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-D-allopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC4
 
 | | Solution Structure of N-Galactosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-D-galactopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC6
 
 | | Solution Structure of N-L-idosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-L-idopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
7EWH
 
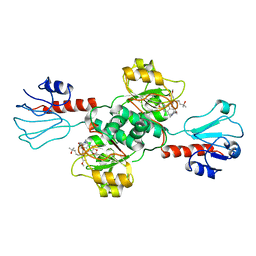 | | Crystal structure of human PHGDH in complex with Homoharringtonine | | 分子名称: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, D-3-phosphoglycerate dehydrogenase | | 著者 | Hsieh, C.H, Cheng, Y.S, Lee, Y.S, Huang, H.C, Juan, H.F. | | 登録日 | 2021-05-25 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Homoharringtonine as a PHGDH inhibitor: Unraveling metabolic dependencies and developing a potent therapeutic strategy for high-risk neuroblastoma.
Biomed Pharmacother, 166, 2023
|
|
4KDF
 
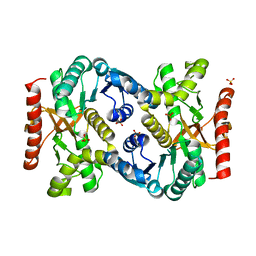 | |
4KDE
 
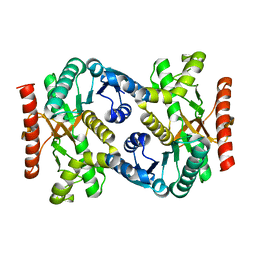 | |
1Z5F
 
 | | Solution Structure of the Cytotoxic RC-RNase 3 with a Pyroglutamate Residue at the N-terminus | | 分子名称: | RC-RNase 3 | | 著者 | Lou, Y.C, Huang, Y.C, Pan, Y.R, Chen, C, Liao, Y.D. | | 登録日 | 2005-03-18 | | 公開日 | 2006-02-28 | | 最終更新日 | 2019-12-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Roles of N-terminal pyroglutamate in maintaining structural integrity and pKa values of catalytic histidine residues in bullfrog ribonuclease 3
J.Mol.Biol., 355, 2006
|
|
7BSL
 
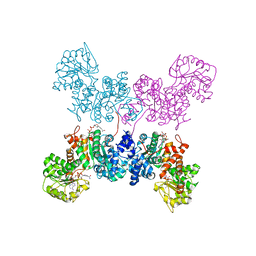 | | Crystal Structure of human ME2 R67A mutant | | 分子名称: | NAD-dependent malic enzyme, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Chen, W.L, Tai, S.C, Hung, H.C, Ho, M.C. | | 登録日 | 2020-03-30 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Single nucleotide variants lead to dysregulation of the human mitochondrial NAD(P) + -dependent malic enzyme.
Iscience, 24, 2021
|
|
7BSJ
 
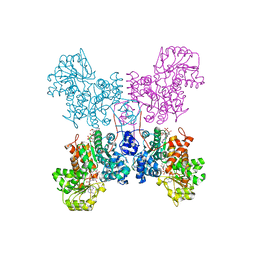 | | Crystal structure of human ME2 R484W | | 分子名称: | FUMARIC ACID, MAGNESIUM ION, NAD-dependent malic enzyme, ... | | 著者 | Chen, W.L, Tai, S.C, Hung, H.C, Ho, M.C. | | 登録日 | 2020-03-30 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Single nucleotide variants lead to dysregulation of the human mitochondrial NAD(P) + -dependent malic enzyme.
Iscience, 24, 2021
|
|
7BSK
 
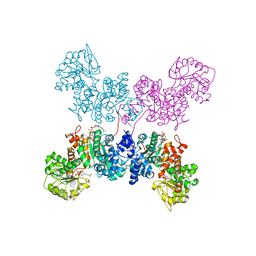 | | Crystal structure of human ME2 R67Q mutant | | 分子名称: | NAD-dependent malic enzyme, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Chen, W.L, Tai, S.C, Hung, H.C, Ho, M.C. | | 登録日 | 2020-03-30 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Single nucleotide variants lead to dysregulation of the human mitochondrial NAD(P) + -dependent malic enzyme.
Iscience, 24, 2021
|
|
