2XPO
 
 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form II | | 分子名称: | CHLORIDE ION, CHROMATIN STRUCTURE MODULATOR, IWS1 | | 著者 | Diebold, M.-L, Koch, M, Cura, V, Moras, D, Cavarelli, J, Romier, C. | | 登録日 | 2010-08-27 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2XPN
 
 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form I | | 分子名称: | BROMIDE ION, CHROMATIN STRUCTURE MODULATOR, IWS1 | | 著者 | Diebold, M.-L, Koch, M, Cura, V, Cavarelli, J, Romier, C. | | 登録日 | 2010-08-27 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2XPP
 
 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form III | | 分子名称: | CHROMATIN STRUCTURE MODULATOR, IWS1 | | 著者 | Diebold, M.-L, Koch, M, Cura, V, Cavarelli, J, Romier, C. | | 登録日 | 2010-08-27 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
5NTC
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0678 | | 分子名称: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | 著者 | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | 登録日 | 2017-04-27 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of mouse CARM1 in complex with inhibitor SA0678
To Be Published
|
|
5IS8
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0271 | | 分子名称: | 1,2-ETHANEDIOL, 5'-{[(3S)-3-amino-3-carboxypropyl](2-chloroethyl)amino}-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, ... | | 著者 | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-03-15 | | 公開日 | 2017-03-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.709 Å) | | 主引用文献 | Crystal structure of mouse CARM1 in complex with inhibitor SA0271
To Be Published
|
|
5G02
 
 | | Crystal Structure of zebrafish Protein Arginine Methyltransferase 2 with SFG | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LITHIUM ION, ... | | 著者 | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-03-16 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.451 Å) | | 主引用文献 | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5FWD
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP2 | | 分子名称: | 1,2-ETHANEDIOL, 9-(7-{[amino(iminio)methyl]amino}-5,6,7-trideoxy-beta-D-ribo-heptofuranosyl)-9H-purin-6-amine, CALCIUM ION, ... | | 著者 | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-02-12 | | 公開日 | 2017-03-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Studies of Protein Arginine Methyltransferase 2 Reveal its Interactions with Potential Substrates and Inhibitors.
FEBS J., 284, 2017
|
|
5FWA
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP1 | | 分子名称: | 1,2-ETHANEDIOL, 9-(6-carbamimidamido-5,6-dideoxy-beta-D-ribo-hexofuranosyl)-9H-purin-6-amine, CALCIUM ION, ... | | 著者 | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-02-16 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5FUB
 
 | | Crystal Structure of zebrafish Protein Arginine Methyltransferase 2 catalytic domain with SAH | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-01-22 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5FUL
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with SAH | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-01-27 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
6TQS
 
 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the conventional FFAT motif of ORP1. | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | 登録日 | 2019-12-17 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6TQU
 
 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the Phospho-FFAT motif of STARD3. | | 分子名称: | Motile sperm domain-containing protein 2, SULFATE ION, StAR-related lipid transfer protein 3 | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | 登録日 | 2019-12-17 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6TQR
 
 | | The crystal structure of the MSP domain of human VAP-A in complex with the Phospho-FFAT motif of STARD3. | | 分子名称: | CHLORIDE ION, StAR-related lipid transfer protein 3, Vesicle-associated membrane protein-associated protein A | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | 登録日 | 2019-12-17 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6TQT
 
 | | The crystal structure of the MSP domain of human MOSPD2. | | 分子名称: | 1,2-ETHANEDIOL, Motile sperm domain-containing protein 2, PHOSPHATE ION | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | 登録日 | 2019-12-17 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
2XP1
 
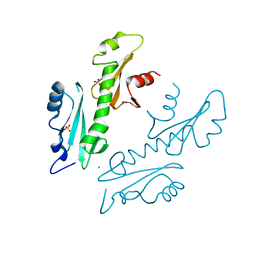 | | Structure of the tandem SH2 domains from Antonospora locustae transcription elongation factor Spt6 | | 分子名称: | CHLORIDE ION, SPT6, SULFATE ION | | 著者 | Diebold, M.-L, Koch, M, Cavarelli, J, Romier, C. | | 登録日 | 2010-08-24 | | 公開日 | 2010-09-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Noncanonical Tandem Sh2 Enables Interaction of Elongation Factor Spt6 with RNA Polymerase II.
J.Biol.Chem., 285, 2010
|
|
5LGS
 
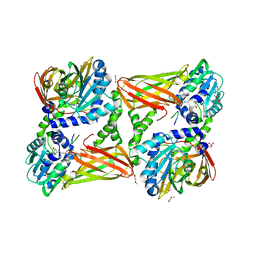 | | Crystal structure of mouse CARM1 in complex with ligand P2C3u | | 分子名称: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | 著者 | Marechal, N, Troffer-Charlier, N, Cura, V, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-07-08 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LGR
 
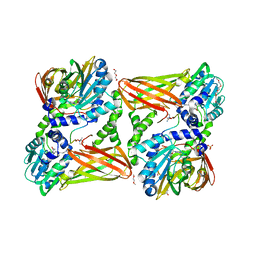 | | Crystal structure of mouse CARM1 in complex with ligand P1C3u | | 分子名称: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | 著者 | Marechal, N, Troffer-Charlier, N, Cura, V, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-07-08 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LV2
 
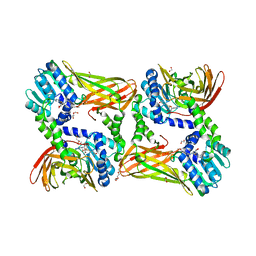 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1246 | | 分子名称: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | 著者 | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-09-12 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
5MFT
 
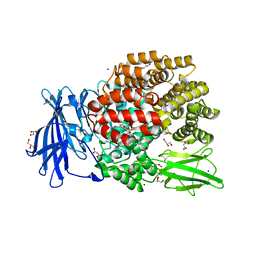 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-1-bromo-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | 分子名称: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | 登録日 | 2016-11-18 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
5MFR
 
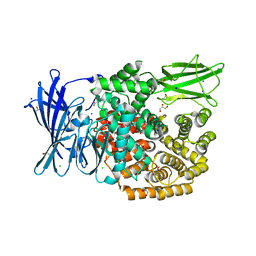 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | 分子名称: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | 登録日 | 2016-11-18 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
5MFS
 
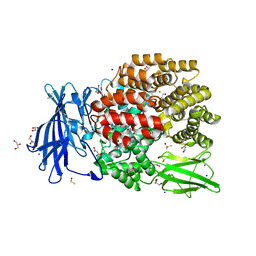 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | 分子名称: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | 登録日 | 2016-11-18 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
5LV5
 
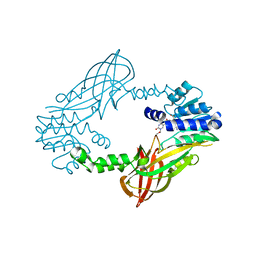 | | Crystal structure of mouse PRMT6 in complex with inhibitor LH1458 | | 分子名称: | 2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]-[(3~{S})-3-azanyl-4-oxidanyl-4-oxidanylidene-butyl]azanium, Protein arginine N-methyltransferase 6 | | 著者 | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-09-12 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
5FQO
 
 | |
5FQN
 
 | |
6SQK
 
 | |
