8BUM
 
 | | Structure of DDB1 bound to DS15-engaged CDK12-cyclin K | | 分子名称: | (2R)-2-[[6-(5-naphthalen-1-ylpentylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU7
 
 | | Structure of DDB1 bound to 21195-engaged CDK12-cyclin K | | 分子名称: | 1,2-ETHANEDIOL, 1-[2,6-bis(chloranyl)phenyl]-6-[[4-(2-hydroxyethyloxy)phenyl]methyl]-3-propan-2-yl-5H-pyrazolo[3,4-d]pyrimidin-4-one, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.245 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUN
 
 | | Structure of DDB1 bound to DS16-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[(4-phenylphenyl)methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU2
 
 | | Structure of DDB1 bound to DS18-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUI
 
 | | Structure of DDB1 bound to DRF-053-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)amino]purin-2-yl]amino]butan-1-ol, 1,2-ETHANEDIOL, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUA
 
 | | Structure of DDB1 bound to 919278-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-~{N}-(1~{H}-benzimidazol-2-yl)-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)propanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.193 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUR
 
 | | Structure of DDB1 bound to DS50-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
6SDW
 
 | |
8P82
 
 | | Cryo-EM structure of dimeric UBR5 | | 分子名称: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | 著者 | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-05-31 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
8P83
 
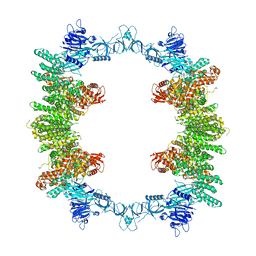 | | Cryo-EM structure of full-length human UBR5 (homotetramer) | | 分子名称: | E3 ubiquitin-protein ligase UBR5 | | 著者 | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-05-31 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
6H0F
 
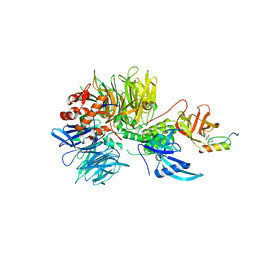 | | Structure of DDB1-CRBN-pomalidomide complex bound to IKZF1(ZF2) | | 分子名称: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, DNA-binding protein Ikaros, Protein cereblon, ... | | 著者 | Petzold, G, Bunker, R.D, Thoma, N.H. | | 登録日 | 2018-07-09 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
6H0G
 
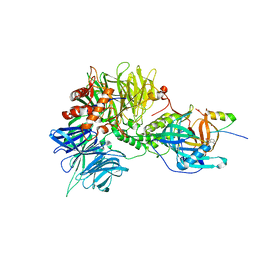 | | Structure of the DDB1-CRBN-pomalidomide complex bound to ZNF692(ZF4) | | 分子名称: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DDB1 (DNA damage binding protein 1),DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | 著者 | Bunker, R.D, Petzold, G, Thoma, N.H. | | 登録日 | 2018-07-09 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (4.25 Å) | | 主引用文献 | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
