5KIY
 
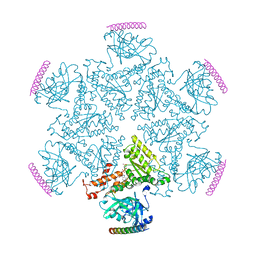 | | p97 ND1-A232E in complex with VIMP | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Selenoprotein S, ... | | 著者 | Tang, W.K, Xia, D. | | 登録日 | 2016-06-17 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural basis for nucleotide-modulated p97 association with the ER membrane.
Cell Discov, 3, 2017
|
|
5KIU
 
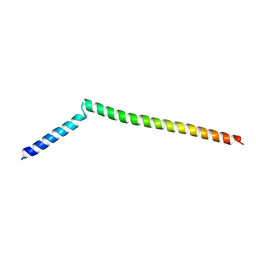 | |
5L7F
 
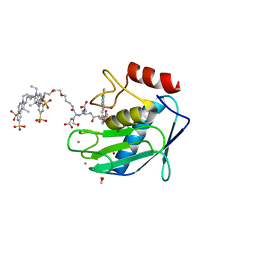 | | Crystal structure of MMP12 mutant K421A in complex with RXP470.1 conjugated with fluorophore Cy5,5 in space group P21. | | 分子名称: | 1,2-ETHANEDIOL, BROMIDE ION, CALCIUM ION, ... | | 著者 | Tepshi, L, Bordenave, T, Rouanet-Mehouas, C, Devel, L, Dive, V, Stura, E.A. | | 登録日 | 2016-06-03 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Synthesis and in Vitro and in Vivo Evaluation of MMP-12 Selective Optical Probes.
Bioconjug.Chem., 27, 2016
|
|
5L79
 
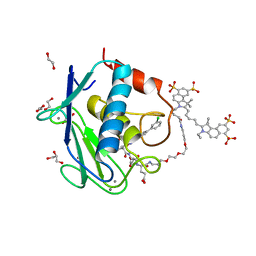 | | Crystal structure of MMP12 in complex with RXP470.1 conjugated with fluorophore Cy5,5 in space group P21212. | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CY5.5-PEG2, ... | | 著者 | Tepshi, L, Bordenave, T, Devel, L, Dive, V, Stura, E.A. | | 登録日 | 2016-06-02 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Synthesis and in Vitro and in Vivo Evaluation of MMP-12 Selective Optical Probes.
Bioconjug.Chem., 27, 2016
|
|
7B3E
 
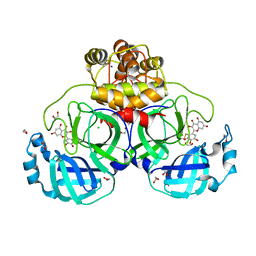 | | Crystal structure of myricetin covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2020-11-30 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7L1G
 
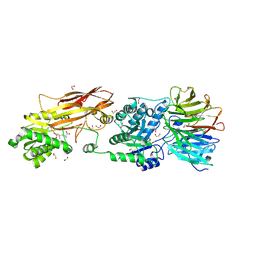 | | PRMT5-MEP50 Complexed with SAM | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Methylosome protein 50, ... | | 著者 | Palte, R.L. | | 登録日 | 2020-12-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Development of a Flexible and Robust Synthesis of Tetrahydrofuro[3,4- b ]furan Nucleoside Analogues.
J.Org.Chem., 86, 2021
|
|
3L59
 
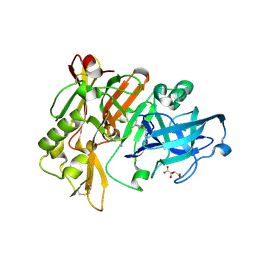 | | Structure of BACE Bound to SCH710413 | | 分子名称: | (2Z)-3-(3-chlorobenzyl)-2-imino-5,5-dimethylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L58
 
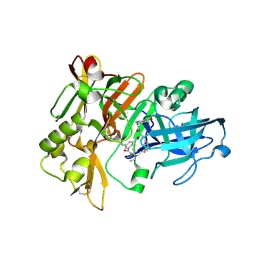 | | Structure of BACE Bound to SCH589432 | | 分子名称: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5B
 
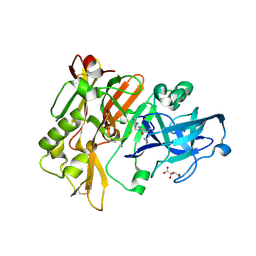 | | Structure of BACE Bound to SCH713601 | | 分子名称: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5E
 
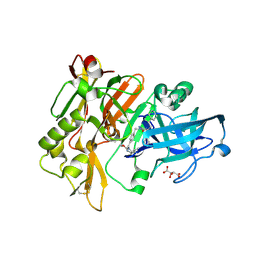 | | Structure of BACE Bound to SCH736062 | | 分子名称: | (4S)-1-(4-{[(2Z,4R)-4-(2-cyclohexylethyl)-4-(cyclohexylmethyl)-2-imino-5-oxoimidazolidin-1-yl]methyl}benzyl)-4-propylimidazolidin-2-one, Beta-secretase 1, D(-)-TARTARIC ACID | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5D
 
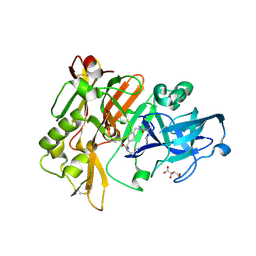 | | Structure of BACE Bound to SCH723873 | | 分子名称: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5F
 
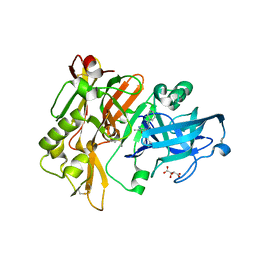 | | Structure of BACE Bound to SCH736201 | | 分子名称: | (2E,5R)-5-(2-cyclohexylethyl)-5-(cyclohexylmethyl)-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5C
 
 | | Structure of BACE Bound to SCH723871 | | 分子名称: | 1-(4-cyanophenyl)-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
4ON3
 
 | | Crystal structure of human sorting nexin 10 (SNX10) | | 分子名称: | NITRATE ION, PENTAETHYLENE GLYCOL, SODIUM ION, ... | | 著者 | Xu, T, Xu, J, Wang, Q, Liu, J. | | 登録日 | 2014-01-28 | | 公開日 | 2014-09-24 | | 最終更新日 | 2014-12-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of human SNX10 reveals insights into its role in human autosomal recessive osteopetrosis.
Proteins, 82, 2014
|
|
4PZG
 
 | | Crystal structure of human sorting nexin 10 (SNX10) | | 分子名称: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, NITRATE ION, Sorting nexin-10 | | 著者 | Xu, T, Xu, J, Wang, Q, Liu, J. | | 登録日 | 2014-03-30 | | 公開日 | 2014-09-24 | | 最終更新日 | 2014-12-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of human SNX10 reveals insights into its role in human autosomal recessive osteopetrosis.
Proteins, 82, 2014
|
|
8I16
 
 | | Crystal structure of the selenomethionine (SeMet)-derived Cas12g (D513A) mutant | | 分子名称: | Cas12g, ZINC ION | | 著者 | Zhang, B, Chen, J, Ye, Y.M, OuYang, S.Y. | | 登録日 | 2023-01-12 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural transitions upon guide RNA binding and their importance in Cas12g-mediated RNA cleavage.
Plos Genet., 19, 2023
|
|
8IN4
 
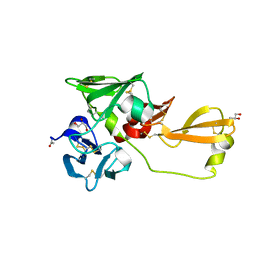 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | 分子名称: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | 著者 | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | 登録日 | 2023-03-08 | | 公開日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN6
 
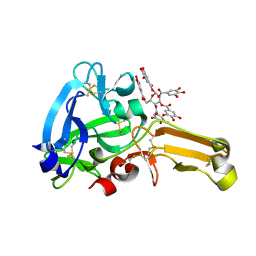 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai complex with tannic acid | | 分子名称: | 25 kDa polyphenol-binding protein, BETA-1,2,3,4,6-PENTA-O-GALLOYL-D-GLUCOPYRANOSE | | 著者 | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | 登録日 | 2023-03-08 | | 公開日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN1
 
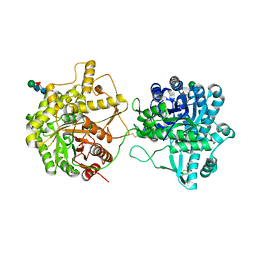 | | beta-glucosidase protein from Aplysia kurodai | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | 登録日 | 2023-03-08 | | 公開日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN3
 
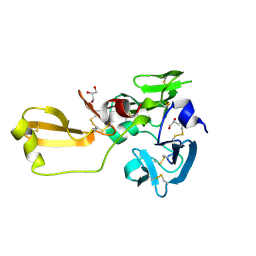 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | 分子名称: | 25 kDa polyphenol-binding protein, GLYCEROL | | 著者 | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | 登録日 | 2023-03-08 | | 公開日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IJK
 
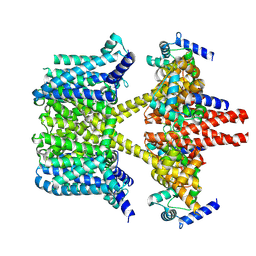 | | human KCNQ2-CaM-Ebio1 complex in the presence of PIP2 | | 分子名称: | Calmodulin-1, N-(1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | 著者 | Ma, D, Guo, J. | | 登録日 | 2023-02-27 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 2024
|
|
5GW0
 
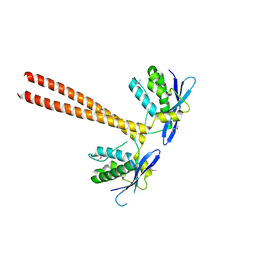 | | Crystal structure of SNX16 PX-Coiled coil | | 分子名称: | Sorting nexin-16 | | 著者 | Xu, J, Liu, J. | | 登録日 | 2016-09-08 | | 公開日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | SNX16 Regulates the Recycling of E-Cadherin through a Unique Mechanism of Coordinated Membrane and Cargo Binding.
Structure, 25, 2017
|
|
5GW1
 
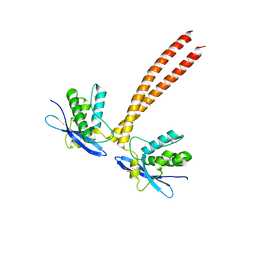 | |
6M11
 
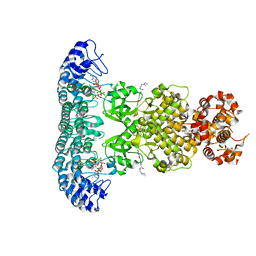 | | Crystal structure of Rnase L in complex with Sunitinib | | 分子名称: | 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, PHOSPHATE ION, ... | | 著者 | Tang, J, Huang, H. | | 登録日 | 2020-02-24 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
6M12
 
 | | Crystal Structure of Rnase L in complex with SU11652 | | 分子名称: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, PHOSPHATE ION, Ribonuclease L, ... | | 著者 | Tang, J, Huang, H. | | 登録日 | 2020-02-24 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
