5ZYL
 
 | | Crystal structure of CERT START domain in complex with compound E25A | | 分子名称: | 2-[4-[4-cyclopentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYI
 
 | | Crystal structure of CERT START domain in complex with compound E16 | | 分子名称: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYJ
 
 | | Crystal structure of CERT START domain in complex with compound E16A | | 分子名称: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYH
 
 | | Crystal structure of CERT START domain in complex with compound E5 | | 分子名称: | 2-[4-[3-~{tert}-butyl-5-[(1~{R},2~{S})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYK
 
 | | Crystal structure of CERT START domain in complex with compound E25 | | 分子名称: | 2-[4-[4-cyclopentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYG
 
 | | Crystal structure of CERT START domain in complex with compound B5 | | 分子名称: | 2-[4-[3-~{tert}-butyl-5-(2-pyridin-2-ylethyl)phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYM
 
 | | Crystal structure of CERT START domain in complex with compound E25B | | 分子名称: | 2-[4-[4-cyclopentyl-3-[(1~{R},2~{S})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
6KKA
 
 | | Xylanase J mutant from Bacillus sp. 41M-1 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Suzuki, M, Takita, T, Nakatani, K. | | 登録日 | 2019-07-24 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
5WR9
 
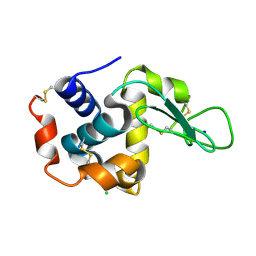 | | Crystal structure of hen egg-white lysozyme | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | 登録日 | 2016-12-01 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5WR8
 
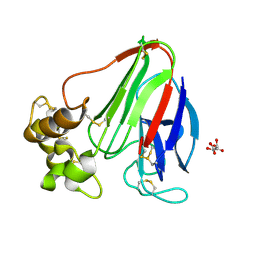 | | Thaumatin structure determined by SACLA at 1.55 Angstrom | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin I | | 著者 | Masuda, T, Suzuki, M, Inoue, S, Sugahara, M. | | 登録日 | 2016-12-01 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
1MP9
 
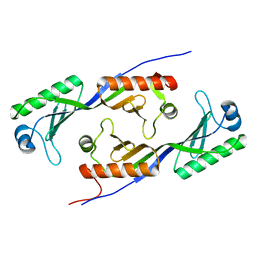 | | TBP from a mesothermophilic archaeon, Sulfolobus acidocaldarius | | 分子名称: | TATA-binding protein | | 著者 | Koike, H, Kawashima-Ohya, Y, Yamasaki, T, Clowney, L, Katsuya, Y, Suzuki, M. | | 登録日 | 2002-09-12 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Origins of Protein Stability Revealed by Comparing Crystal Structures of TATA Binding Proteins.
Structure, 12, 2004
|
|
2GCC
 
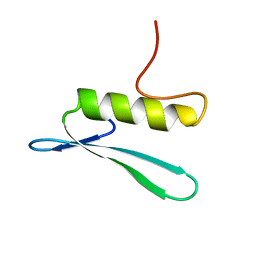 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | 分子名称: | ATERF1 | | 著者 | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | 登録日 | 1998-03-13 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
4W4Q
 
 | | Glucose isomerase structure determined by serial femtosecond crystallography at SACLA | | 分子名称: | CALCIUM ION, Xylose isomerase | | 著者 | Nango, E, Tanaka, T, Sugahara, M, Suzuki, M, Iwata, S. | | 登録日 | 2014-08-15 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat.Methods, 12, 2015
|
|
6LCQ
 
 | | Crystal structure of rice defensin OsAFP1 | | 分子名称: | Defensin-like protein CAL1, PHOSPHATE ION | | 著者 | Ochiai, A, Ogawa, K, Fukuda, M, Suzuki, M, Ito, K, Tanaka, T, Sagehashi, Y, Taniguchi, M. | | 登録日 | 2019-11-19 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystal structure of rice defensin OsAFP1 and molecular insight into lipid-binding.
J.Biosci.Bioeng., 130, 2020
|
|
5KXV
 
 | | Structure Proteinase K at 0.98 Angstroms | | 分子名称: | CALCIUM ION, GLYCEROL, NITRATE ION, ... | | 著者 | Masuda, T, Suzuki, M, Inoue, S, Numata, K, Sugahara, M. | | 登録日 | 2016-07-20 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Atomic resolution structure of serine protease proteinase K at ambient temperature.
Sci Rep, 7, 2017
|
|
3KLR
 
 | | Bovine H-protein at 0.88 angstrom resolution | | 分子名称: | GLYCEROL, Glycine cleavage system H protein, SULFATE ION | | 著者 | Higashiura, A, Kurakane, T, Matsuda, M, Suzuki, M, Inaka, K, Sato, M, Tanaka, H, Fujiwara, K, Nakagawa, A. | | 登録日 | 2009-11-09 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (0.88 Å) | | 主引用文献 | High-resolution X-ray crystal structure of bovine H-protein at 0.88 A resolution
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7D5M
 
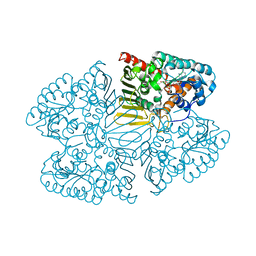 | | Crystal structure of inositol dehydrogenase homolog complexed with NAD+ from Azotobacter vinelandii | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase | | 著者 | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | 登録日 | 2020-09-27 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of inositol dehydrogenase complexed with NAD+ from Azotobacter vinelandii
To Be Published
|
|
7D5N
 
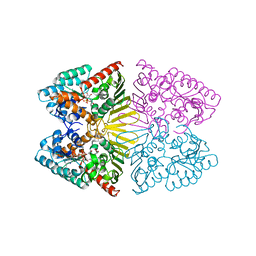 | | Crystal structure of inositol dehydrogenase homolog complexed with NADH and myo-inositol from Azotobacter vinelandii | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Oxidoreductase | | 著者 | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | 登録日 | 2020-09-27 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of inositol dehydrogenase complexed with NADH and myo-inositol from Azotobacter vinelandii
To Be Published
|
|
3VWU
 
 | |
3VWW
 
 | |
6LFE
 
 | | Rat-COMT, Nitecapone,SAM and Mg bond | | 分子名称: | 3-(3,4-dihydroxy-5-nitrobenzylidene)pentane-2,4-dione, Catechol O-methyltransferase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Takebe, K, Iijima, H, Suzuki, M. | | 登録日 | 2019-12-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of Catechol O-Methyltransferase Complexed with Nitecapone.
Chem Pharm Bull (Tokyo), 68, 2020
|
|
6LS1
 
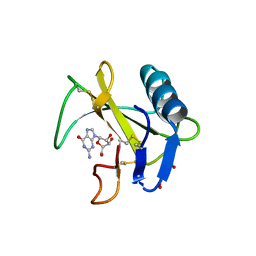 | | Ribonuclease from Hericium erinaceus active and GMP binding form | | 分子名称: | DI(HYDROXYETHYL)ETHER, GUANOSINE, Ribonuclease T1, ... | | 著者 | Takebe, K, Suzuki, M, Sangawa, T, Kobayashi, H, Itagaki, T. | | 登録日 | 2020-01-16 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Ribonuclease from Hericium erinaceus active and GMP binding form
To Be Published
|
|
5KXU
 
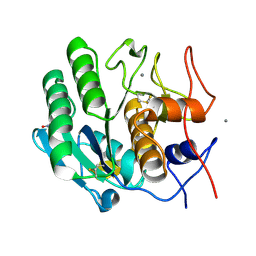 | | Structure Proteinase K determined by SACLA | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Masuda, T, Suzuki, M, Inoue, S, Numata, K, Sugahara, M. | | 登録日 | 2016-07-20 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Atomic resolution structure of serine protease proteinase K at ambient temperature.
Sci Rep, 7, 2017
|
|
3VU9
 
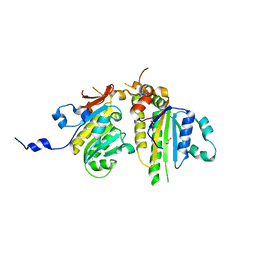 | | Crystal Structure of Psy3-Csm2 complex | | 分子名称: | 1,2-ETHANEDIOL, Chromosome segregation in meiosis protein 2, Platinum sensitivity protein 3 | | 著者 | Tawaramoto, M, Sasanuma, H, Hosaka, H, Lao, J.P, Sanda, E, Suzuki, M, Yamashita, E, Hunter, N, Shinohara, M, Nakagawa, A, Shinohara, A. | | 登録日 | 2012-06-23 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A new protein complex promoting the assembly of Rad51 filaments
Nat Commun, 4, 2013
|
|
1O3Y
 
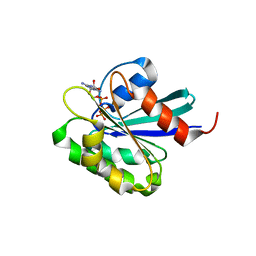 | | Crystal structure of mouse ARF1 (delta17-Q71L), GTP form | | 分子名称: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | 著者 | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | 登録日 | 2003-05-08 | | 公開日 | 2003-05-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
Nat.Struct.Biol., 10, 2003
|
|
