8SQJ
 
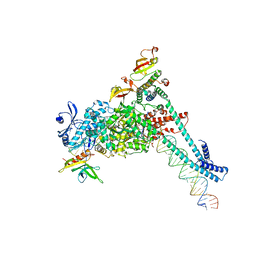 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode | | 分子名称: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | 著者 | Small, G.I, Darst, S.A, Campbell, E.A. | | 登録日 | 2023-05-04 | | 公開日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
6ZJV
 
 | |
6ZJP
 
 | |
6ZJW
 
 | |
6ZJX
 
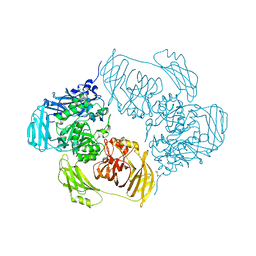 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with saccharose | | 分子名称: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
3KWV
 
 | |
7Q7Y
 
 | |
7Q82
 
 | |
8E9E
 
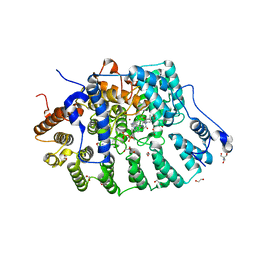 | | Rat protein farnesyltransferase in complex with FPP and inhibitor 2f | | 分子名称: | (5S)-4-({1-[(4-bromophenyl)methyl]-1H-imidazol-5-yl}methyl)-5-butyl-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Wang, Y, Shi, Y, Beese, L.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.844 Å) | | 主引用文献 | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
6S4Q
 
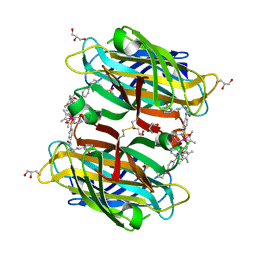 | | scdSav(SASK) - Engineering Single-Chain Dimeric Streptavidin as Host for Artificial Metalloenzymes | | 分子名称: | GLYCEROL, Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | 著者 | Rebelein, J.G. | | 登録日 | 2019-06-28 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Breaking Symmetry: Engineering Single-Chain Dimeric Streptavidin as Host for Artificial Metalloenzymes.
J.Am.Chem.Soc., 141, 2019
|
|
6S50
 
 | |
7Q7Z
 
 | |
7Q81
 
 | |
7Q7X
 
 | |
7Q80
 
 | |
6SE8
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Beta-galactosidase, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.835 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6SEA
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactose bound in deep mode | | 分子名称: | ACETATE ION, Beta-galactosidase, SODIUM ION, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.869 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6SE9
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactose bound in shallow mode | | 分子名称: | ACETATE ION, Beta-galactosidase, FORMIC ACID, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.965 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6H1P
 
 | |
6SED
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with galactose | | 分子名称: | ACETATE ION, Beta-galactosidase, FORMIC ACID, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.233 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6SEB
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG | | 分子名称: | 1-methylethyl 1-thio-beta-D-galactopyranoside, ACETATE ION, Beta-galactosidase, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.272 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
1TFR
 
 | | RNASE H FROM BACTERIOPHAGE T4 | | 分子名称: | MAGNESIUM ION, T4 RNASE H | | 著者 | Mueser, T.C, Nossal, N.G, Hyde, C.C. | | 登録日 | 1996-04-27 | | 公開日 | 1996-11-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Structure of bacteriophage T4 RNase H, a 5' to 3' RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins.
Cell(Cambridge,Mass.), 85, 1996
|
|
5ONI
 
 | | LOW-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | 分子名称: | 1,4-BUTANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | 著者 | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | 登録日 | 2017-08-03 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
5DZO
 
 | |
5CSE
 
 | | Streptavidin-S112Y-K121E Complexed with Palladium-Containing Biotin Ligand | | 分子名称: | CHLORIDE ION, Streptavidin, chloro{di-tert-butyl[2-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)ethyl]-lambda~5~-phosphanyl}(1-phenylprop-1-ene-1,3-diyl-kappa~2~C~1~,C~3~)palladium | | 著者 | Finke, A.D, Vera, L, Marsh, M, Chatterjee, A, Ward, T.R. | | 登録日 | 2015-07-23 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | An enantioselective artificial Suzukiase based on the biotin-streptavidin technology.
Chem Sci, 7, 2016
|
|
