5Y1Y
 
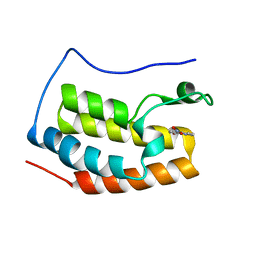 | |
7WIJ
 
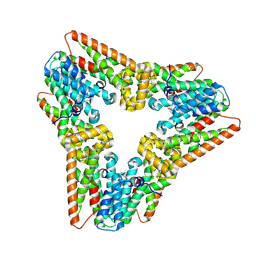 | |
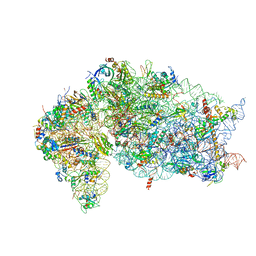
7JQC
 
 | | SARS-CoV-2 Nsp1, CrPV IRES and rabbit 40S ribosome complex | | 分子名称: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | 著者 | Yuan, S, Xiong, Y. | | 登録日 | 2020-08-10 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
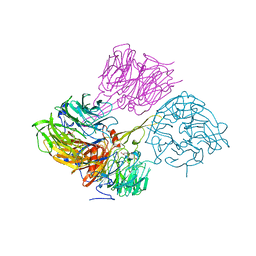
7JQB
 
 | | SARS-CoV-2 Nsp1 and rabbit 40S ribosome complex | | 分子名称: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | 著者 | Yuan, S, Xiong, Y. | | 登録日 | 2020-08-10 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | 分子名称: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | 著者 | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | 登録日 | 2019-12-18 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.033 Å) | | 主引用文献 | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
4MH1
 
 | |
4IOP
 
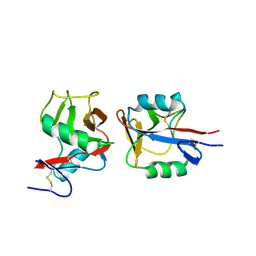 | | Crystal structure of NKp65 bound to its ligand KACL | | 分子名称: | C-type lectin domain family 2 member A, Killer cell lectin-like receptor subfamily F member 2, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Li, Y. | | 登録日 | 2013-01-08 | | 公開日 | 2013-07-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of NKp65 bound to its keratinocyte ligand reveals basis for genetically linked recognition in natural killer gene complex.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
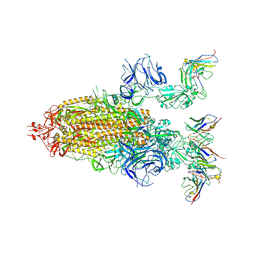
8DZI
 
 | |
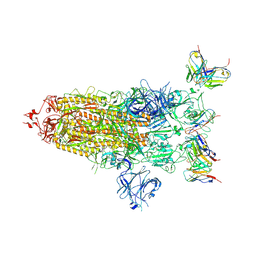
8DZH
 
 | |
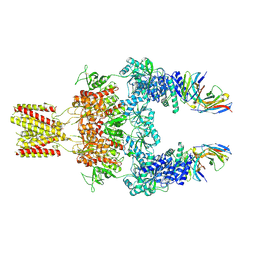
8JJ1
 
 | |
8JIZ
 
 | |
8JJ2
 
 | |
8JJ0
 
 | |
8KGK
 
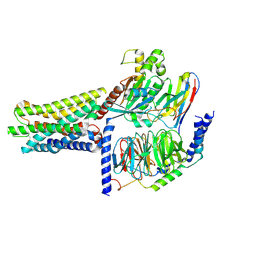 | | Cryo-EM structure of the GPR61-Gs complex | | 分子名称: | G-protein coupled receptor 61, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Nie, Y, Qiu, Z, Zheng, S. | | 登録日 | 2023-08-19 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G s signaling.
Nat Commun, 14, 2023
|
|
7L5D
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with demethylated analog of masitinib | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-12-21 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
8IVZ
 
 | | Crystal structure of talin R7 in complex with KANK1 KN motif | | 分子名称: | KN motif and ankyrin repeat domains 1, Talin-1 | | 著者 | Xu, Y, Li, K, Wei, Z, Cong, Y. | | 登録日 | 2023-03-29 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
4RAP
 
 | | Crystal structure of bacterial iron-containing dodecameric glycosyltransferase TibC from enterotoxigenic E.coli H10407 | | 分子名称: | 1,2-ETHANEDIOL, FE (III) ION, Glycosyltransferase TibC | | 著者 | Yao, Q, Lu, Q, Xu, Y, Shao, F. | | 登録日 | 2014-09-10 | | 公開日 | 2014-10-29 | | 実験手法 | X-RAY DIFFRACTION (2.881 Å) | | 主引用文献 | A structural mechanism for bacterial autotransporter glycosylation by a dodecameric heptosyltransferase family.
Elife, 3, 2014
|
|
8KB0
 
 | | Crystal structure of 01JD-AEAIIVAMV | | 分子名称: | Beta2-microglobulin, MHC class I antigen alpha chain, peptide of AIV | | 著者 | Tang, Z, Zhang, N. | | 登録日 | 2023-08-03 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
8KB1
 
 | | Crystal structure of 11JD | | 分子名称: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | 著者 | Tang, Z, Zhang, N. | | 登録日 | 2023-08-03 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
2D2D
 
 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor I2 | | 分子名称: | 3C-like proteinase, ETHYL (2E,4S)-4-[((2R)-2-{[N-(TERT-BUTOXYCARBONYL)-L-VALYL]AMINO}-2-PHENYLETHANOYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENT-2-ENOATE | | 著者 | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Ding, Y, Rao, Z. | | 登録日 | 2005-09-08 | | 公開日 | 2005-09-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
5Z2T
 
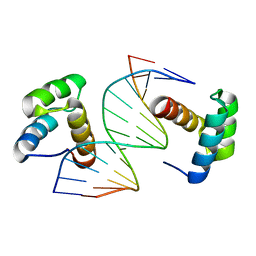 | | Crystal structure of DNA-bound DUX4-HD2 | | 分子名称: | 5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*TP*T)-3', 5'-D(P*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3', Double homeobox protein 4 | | 著者 | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | 登録日 | 2018-01-04 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.623 Å) | | 主引用文献 | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
8HXQ
 
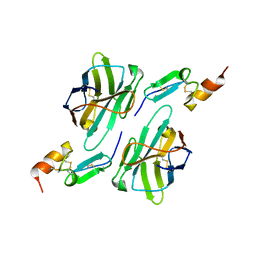 | | Nanobody1 in complex with human BCMA ECD | | 分子名称: | Nanobody1, Tumor necrosis factor receptor superfamily member 17 | | 著者 | Sun, Y, Zhang, B. | | 登録日 | 2023-01-05 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther, 8, 2023
|
|
8HXR
 
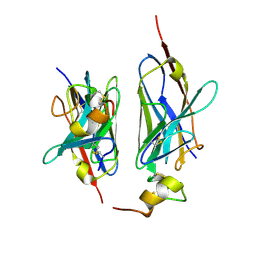 | | Nanobody2 in complex with human BCMA ECD | | 分子名称: | Nanobody2, Tumor necrosis factor receptor superfamily member 17 | | 著者 | Sun, Y, Zhang, B. | | 登録日 | 2023-01-05 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther, 8, 2023
|
|
7E3O
 
 | |
8KG5
 
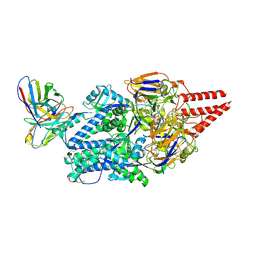 | | Prefusion RSV F Bound to Lonafarnib and D25 Fab | | 分子名称: | 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, D25 heavy chain, D25 light chain, ... | | 著者 | Yang, Q, Xue, B, Liu, F, Peng, W, Chen, X. | | 登録日 | 2023-08-17 | | 公開日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Farnesyltransferase inhibitor lonafarnib suppresses respiratory syncytial virus infection by blocking conformational change of fusion glycoprotein.
Signal Transduct Target Ther, 9, 2024
|
|
