7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-08-31 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
3FF7
 
 | | Structure of NK cell receptor KLRG1 bound to E-cadherin | | 分子名称: | ACETIC ACID, Epithelial cadherin, Killer cell lectin-like receptor subfamily G member 1 | | 著者 | Li, Y, Mariuzza, R.A. | | 登録日 | 2008-12-02 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
3FF9
 
 | | Structure of NK cell receptor KLRG1 | | 分子名称: | Killer cell lectin-like receptor subfamily G member 1 | | 著者 | Li, Y, Mariuzza, R.A. | | 登録日 | 2008-12-02 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
7WTK
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv286 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv286, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTJ
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv286 | | 分子名称: | Heavy chain of XGv286, Light chain of XGv286, Spike protein S1 | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTG
 
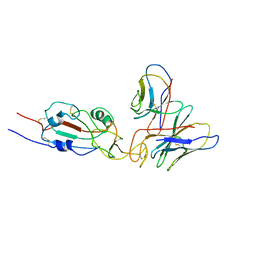 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv051 | | 分子名称: | Heavy chain of XGv051, Light chain of XGv051, Spike protein S1 | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTH
 
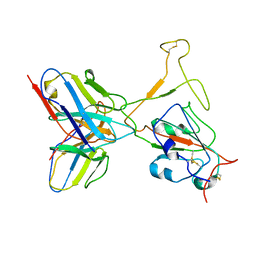 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv264 | | 分子名称: | Heavy chain of XGv264, Light chain of XGv264, Spike protein S1 | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTF
 
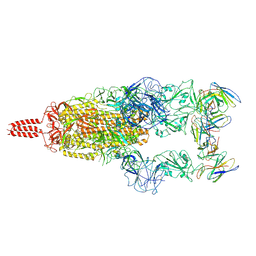 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv051 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv051, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTI
 
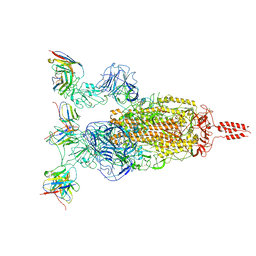 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv264 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv264, Light chain of XGv264, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-21 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
5WWL
 
 | | Crystal structure of the Schizogenesis pombe kinetochore Mis12C subcomplex | | 分子名称: | Centromere protein mis12, Kinetochore protein nnf1 | | 著者 | Wang, C, Zhou, X, Wu, M, Zhang, X, Zang, J. | | 登録日 | 2017-01-02 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3FF8
 
 | | Structure of NK cell receptor KLRG1 bound to E-cadherin | | 分子名称: | CALCIUM ION, Epithelial cadherin, Killer cell lectin-like receptor subfamily G member 1 | | 著者 | Li, Y, Mariuzza, R.A. | | 登録日 | 2008-12-02 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
7CHF
 
 | |
7CHH
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-368-2 Fab heavy chain, ... | | 著者 | Xiao, J, Zhu, Q, Wang, G. | | 登録日 | 2020-07-05 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
7CHE
 
 | |
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | 著者 | Xiao, J, Zhu, Q. | | 登録日 | 2020-07-05 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
4TY9
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | 分子名称: | 5-(trifluoromethyl)pyridin-2-amine, Polyprotein | | 著者 | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | 登録日 | 2014-07-08 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYB
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | 分子名称: | (2R)-morpholin-4-yl(phenyl)ethanenitrile, Polyprotein | | 著者 | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | 登録日 | 2014-07-08 | | 公開日 | 2015-05-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYA
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | 分子名称: | 4-(trifluoromethyl)benzoic acid, Polyprotein | | 著者 | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | 登録日 | 2014-07-08 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
5GI0
 
 | | Crystal structure of RNA editing factor MORF9/RIP9 | | 分子名称: | Multiple organellar RNA editing factor 9, chloroplastic | | 著者 | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | 登録日 | 2016-06-21 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.044 Å) | | 主引用文献 | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
7CHC
 
 | |
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | 分子名称: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | 著者 | Guo, Y, Wang, S, Nie, Y, Li, S. | | 登録日 | 2018-06-21 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
1LQF
 
 | | Structure of PTP1b in Complex with a Peptidic Bisphosphonate Inhibitor | | 分子名称: | N-BENZOYL-L-GLUTAMYL-[4-PHOSPHONO(DIFLUOROMETHYL)]-L-PHENYLALANINE-[4-PHOSPHONO(DIFLUORO-METHYL)]-L-PHENYLALANINEAMIDE, protein-tyrosine phosphatase, non-receptor type 1 | | 著者 | Asante-Appiah, E, Patel, S, Dufresne, C, Scapin, G. | | 登録日 | 2002-05-10 | | 公開日 | 2002-07-24 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The structure of PTP-1B in complex with a peptide inhibitor reveals an alternative binding mode for bisphosphonates.
Biochemistry, 41, 2002
|
|
9J4X
 
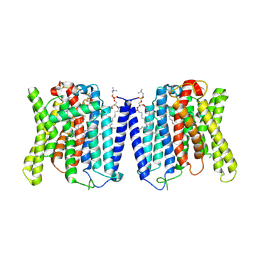 | | CryoEM structure of human XPR1 in apo state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Solute carrier family 53 member 1 | | 著者 | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | 登録日 | 2024-08-10 | | 公開日 | 2024-12-04 | | 最終更新日 | 2025-01-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
