2FBW
 
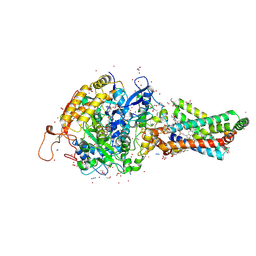 | | Avian respiratory complex II with carboxin bound | | 分子名称: | (~{Z})-2-oxidanylbut-2-enedioic acid, 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, AZIDE ION, ... | | 著者 | Huang, L.S, Sun, G, Cobessi, D, Wang, A.C, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | 登録日 | 2005-12-10 | | 公開日 | 2005-12-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme.
J.Biol.Chem., 281, 2006
|
|
2H89
 
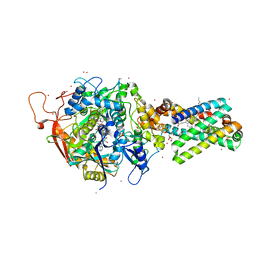 | | Avian Respiratory Complex II with Malonate Bound | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | 著者 | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | 登録日 | 2006-06-06 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
6MYP
 
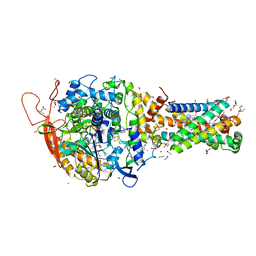 | | Avian mitochondrial complex II with TTFA (thenoyltrifluoroacetone) bound | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 4,4,4-TRIFLUORO-1-THIEN-2-YLBUTANE-1,3-DIONE, BICARBONATE ION, ... | | 著者 | Berry, E.A, Huang, L.-S. | | 登録日 | 2018-11-02 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYQ
 
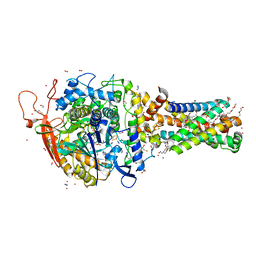 | | Avian mitochondrial complex II with ferulenol bound | | 分子名称: | (~{Z})-2-oxidanylbut-2-enedioic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, ... | | 著者 | Berry, E.A, Huang, L.-S. | | 登録日 | 2018-11-02 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYT
 
 | | Avian mitochondrial complex II with Atpenin A5 bound, sidechain in pocket | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Berry, E.A, Huang, L.-S. | | 登録日 | 2018-11-02 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYO
 
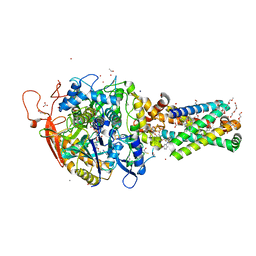 | | Avian mitochondrial complex II with flutolanyl bound | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Berry, E.A, Huang, L.-S. | | 登録日 | 2018-11-02 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
8GPY
 
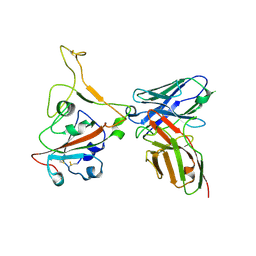 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | 分子名称: | Spike protein S1, scFv | | 著者 | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | 登録日 | 2022-08-27 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
6MYU
 
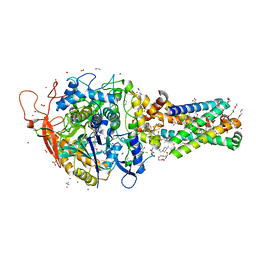 | |
2H88
 
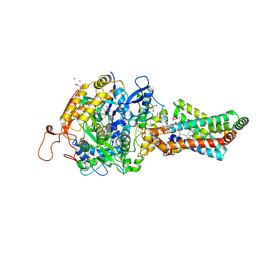 | | Avian Mitochondrial Respiratory Complex II at 1.8 Angstrom Resolution | | 分子名称: | AZIDE ION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | 著者 | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | 登録日 | 2006-06-06 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
1A6S
 
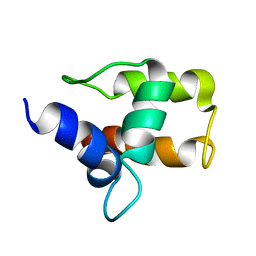 | | M-DOMAIN FROM GAG POLYPROTEIN OF ROUS SARCOMA VIRUS, NMR, 20 STRUCTURES | | 分子名称: | GAG POLYPROTEIN | | 著者 | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Zhou, W, Wolven, A, Wilson, C.B, Nelle, T.D, Resh, M.D, Wills, J, Cowburn, D. | | 登録日 | 1998-03-02 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus
J.Mol.Biol., 279, 1998
|
|
5F22
 
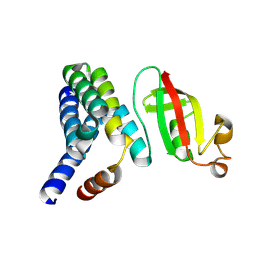 | |
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | 分子名称: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | 登録日 | 2022-04-19 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
6JYT
 
 | |
7D3M
 
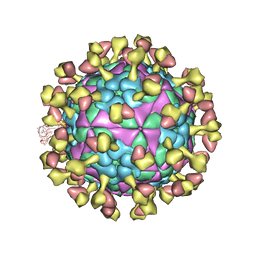 | |
7D3K
 
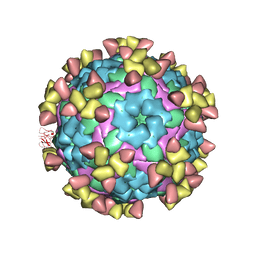 | |
7D3R
 
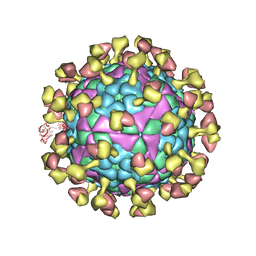 | |
7D3L
 
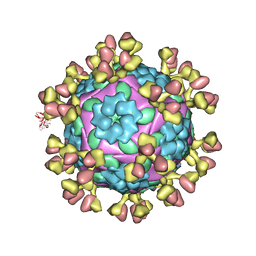 | |
6J17
 
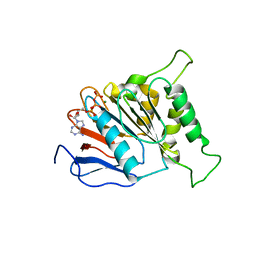 | | ATPase | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ESX-3 secretion system protein EccC3, MAGNESIUM ION | | 著者 | Wang, S.H, Li, J, Rao, Z.H. | | 登録日 | 2018-12-28 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.975 Å) | | 主引用文献 | Structural insights into substrate recognition by the type VII secretion system.
Protein Cell, 11, 2020
|
|
7BX8
 
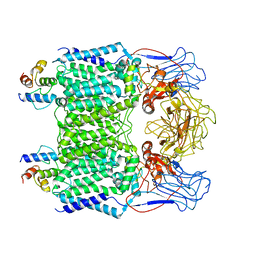 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric "resting state" | | 分子名称: | Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein | | 著者 | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | 登録日 | 2020-04-17 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7BOK
 
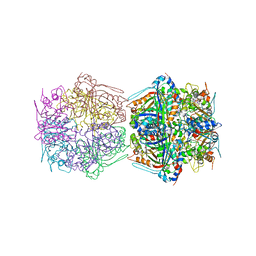 | | Cryo-EM structure of the encapsulated DyP-type peroxidase from Mycobacterium smegmatis | | 分子名称: | Dyp-type peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BOJ
 
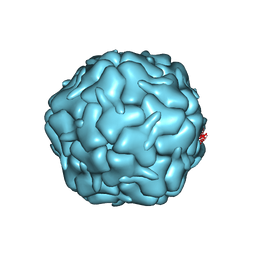 | | Cryo-EM structure of the encapsulin shell from Mycobacterium smegmatis | | 分子名称: | 29 kDa antigen Cfp29 | | 著者 | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6J72
 
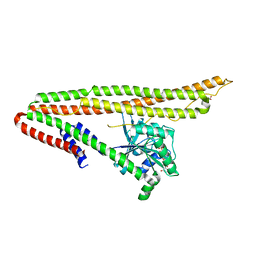 | | Crystal structure of IniA from Mycobacterium smegmatis with GTP bound | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Isoniazid inducible gene protein IniA, L(+)-TARTARIC ACID, ... | | 著者 | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | 登録日 | 2019-01-16 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
7X6W
 
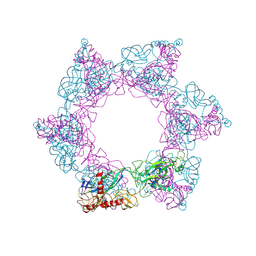 | | SFTSV 2 fold hexamer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Sun, Z, Lou, Z. | | 登録日 | 2022-03-08 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (5.18 Å) | | 主引用文献 | Architecture of severe fever with thrombocytopenia syndrome virus.
Protein Cell, 14, 2023
|
|
7X6U
 
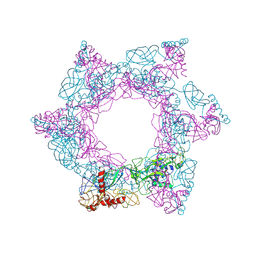 | | SFTSV 3 fold hexmer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Sun, Z, Lou, Z. | | 登録日 | 2022-03-08 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Architecture of severe fever with thrombocytopenia syndrome virus.
Protein Cell, 14, 2023
|
|
7X72
 
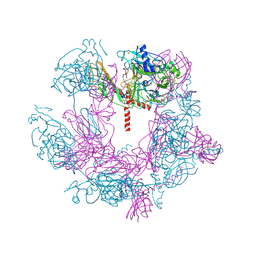 | | SFTSV 5 fold pentamer | | 分子名称: | Envelopment polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Sun, Z, Lou, Z. | | 登録日 | 2022-03-08 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Architecture of severe fever with thrombocytopenia syndrome virus.
Protein Cell, 14, 2023
|
|
