1WQW
 
 | |
1WN2
 
 | |
1WQ7
 
 | |
1WPY
 
 | |
5DGQ
 
 | |
6JZH
 
 | | Structure of human A2A adenosine receptor in complex with ZM241385 obtained from SFX experiments under atmospheric pressure | | 分子名称: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | 著者 | Nango, E, Shimamura, T, Nakane, T, Yamanaka, Y, Mori, C, Kimura, K.T, Fujiwara, T, Tanaka, T, Iwata, S. | | 登録日 | 2019-05-02 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
J.Appl.Crystallogr., 52, 2019
|
|
5DGR
 
 | | Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose, Putative endoglucanase-related protein, SODIUM ION | | 著者 | Suzuki, K, Honda, Y, Fushinobu, S. | | 登録日 | 2015-08-28 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of an inverting glycoside hydrolase family 9 exo-beta-D-glucosaminidase and the design of glycosynthase.
Biochem.J., 473, 2016
|
|
3QXM
 
 | | Crystal Structure of Human GluK2 Ligand-Binding Core in Complex with Novel Marine-Derived Toxins, Neodysiherbaine A | | 分子名称: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, Glutamate receptor ionotropic, kainate 2 | | 著者 | Unno, M, Sasaki, M, Ikeda-Saito, M. | | 登録日 | 2011-03-02 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
4JAW
 
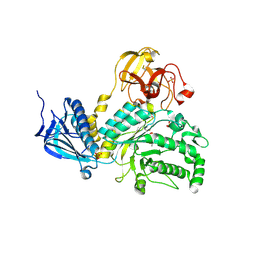 | | Crystal Structure of Lacto-N-Biosidase from Bifidobacterium bifidum complexed with LNB-thiazoline | | 分子名称: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Lacto-N-biosidase, SULFATE ION, ... | | 著者 | Ito, T, Katayama, T, Stubbs, K.A, Fushinobu, S. | | 登録日 | 2013-02-19 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum
J.Biol.Chem., 288, 2013
|
|
2G87
 
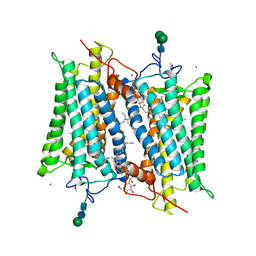 | | Crystallographic model of bathorhodopsin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | 著者 | Nakamichi, H, Okada, T. | | 登録日 | 2006-03-02 | | 公開日 | 2006-09-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystallographic analysis of primary visual photochemistry
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
4H04
 
 | | Lacto-N-biosidase from Bifidobacterium bifidum | | 分子名称: | Lacto-N-biosidase, SULFATE ION, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Ito, T, Katayama, T, Wada, J, Suzuki, R, Ashida, H, Wakagi, T, Yamamoto, K, Fushinobu, S. | | 登録日 | 2012-09-07 | | 公開日 | 2013-03-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum
J.Biol.Chem., 288, 2013
|
|
1EHL
 
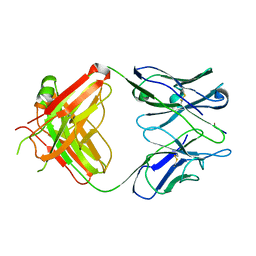 | | 64M-2 ANTIBODY FAB COMPLEXED WITH D(5HT)(6-4)T | | 分子名称: | 5'-(D(5HT)P*(6-4)T)-3', ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (HEAVY CHAIN), ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (LIGHT CHAIN) | | 著者 | Yokoyama, H, Mizutani, R, Satow, Y, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | 登録日 | 2000-02-21 | | 公開日 | 2001-02-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the 64M-2 antibody Fab fragment in complex with a DNA dT(6-4)T photoproduct formed by ultraviolet radiation.
J.Mol.Biol., 299, 2000
|
|
2DDX
 
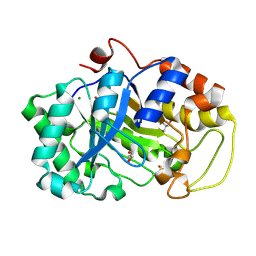 | |
1KEG
 
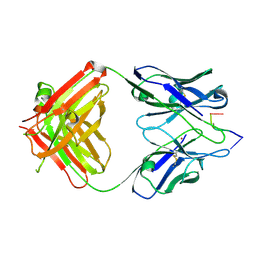 | | Antibody 64M-2 Fab complexed with dTT(6-4)TT | | 分子名称: | 5'-D(*TP*(64T)P*TP*T)-3', Anti-(6-4) photoproduct antibody 64M-2 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-2 Fab (light chain), ... | | 著者 | Yokoyama, H, Mizutani, R, Satow, Y, Sato, K, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | 登録日 | 2001-11-15 | | 公開日 | 2002-11-15 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of the DNA (6-4) photoproduct dTT(6-4)TT in complex with the 64M-2 antibody Fab fragment implies increased antibody-binding affinity by the flanking nucleotides.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5GU7
 
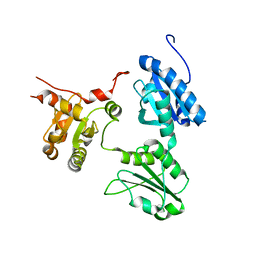 | | Crystal Structure of human ERp44 form II | | 分子名称: | Endoplasmic reticulum resident protein 44 | | 著者 | Watanabe, S, Inaba, K. | | 登録日 | 2016-08-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5GU6
 
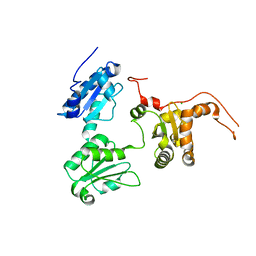 | | Crystal structure of Human ERp44 form I | | 分子名称: | CHLORIDE ION, Endoplasmic reticulum resident protein 44 | | 著者 | Watanabe, S, Inaba, K. | | 登録日 | 2016-08-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3FV1
 
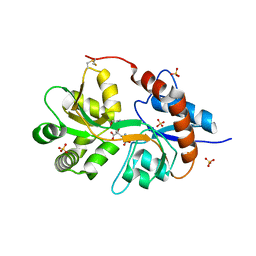 | | Crystal Structure of the human glutamate receptor, GluR5, ligand-binding core in complex with dysiherbaine in space group P1 | | 分子名称: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | 著者 | Unno, M, Sasaki, M, Ikeda-Saito, M. | | 登録日 | 2009-01-15 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FVG
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with MSVIII-19 in space group P1 | | 分子名称: | (2R,3aR,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | 著者 | Unno, M, Sasaki, M, Ikeda-Saito, M. | | 登録日 | 2009-01-15 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
1SYV
 
 | | HLA-B*4405 complexed to the dominant self ligand EEFGRAYGF | | 分子名称: | Beta-2-microglobulin, MHC class I antigen, major histocompatibility complex, ... | | 著者 | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | 登録日 | 2004-04-02 | | 公開日 | 2004-10-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
1SYS
 
 | | Crystal structure of HLA, B*4403, and peptide EEPTVIKKY | | 分子名称: | Beta-2-microglobulin, Sorting nexin 5, leukocyte antigen (HLA) class I molecule | | 著者 | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | 登録日 | 2004-04-01 | | 公開日 | 2004-10-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
2EYT
 
 | |
2EYS
 
 | |
2EYR
 
 | |
4QRP
 
 | | Crystal Structure of HLA B*0801 in complex with HSKKKCDEL and DD31 TCR | | 分子名称: | Beta-2-microglobulin, DD31 TCR alpha chain, DD31 TCR beta chain, ... | | 著者 | Gras, S, Berry, R, Lucet, I.S, Rossjohn, J. | | 登録日 | 2014-07-02 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | An Extensive Antigenic Footprint Underpins Immunodominant TCR Adaptability against a Hypervariable Viral Determinant.
J.Immunol., 193, 2014
|
|
4QRQ
 
 | | Crystal Structure of HLA B*0801 in complex with HSKKKCDEL | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | 著者 | Gras, S, Berry, R, Lucet, I.S, Rossjohn, J. | | 登録日 | 2014-07-02 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | An Extensive Antigenic Footprint Underpins Immunodominant TCR Adaptability against a Hypervariable Viral Determinant.
J.Immunol., 193, 2014
|
|
