1LWB
 
 | |
6K3N
 
 | |
1C52
 
 | | THERMUS THERMOPHILUS CYTOCHROME-C552: A NEW HIGHLY THERMOSTABLE CYTOCHROME-C STRUCTURE OBTAINED BY MAD PHASING | | 分子名称: | CYTOCHROME-C552, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Than, M.E, Hof, P, Huber, R, Bourenkov, G.P, Bartunik, H.D, Buse, G, Soulimane, T. | | 登録日 | 1997-06-23 | | 公開日 | 1998-06-24 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing.
J.Mol.Biol., 271, 1997
|
|
6LZP
 
 | |
7MCI
 
 | | MoFe protein from Azotobacter vinelandii with a sulfur-replenished cofactor | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Kang, W, Lee, C, Hu, Y, Ribbe, M.W. | | 登録日 | 2021-04-02 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Evidence of substrate binding and product release via belt-sulfur mobilization of the nitrogenase cofactor
Nat Catal, 5, 2022
|
|
2GLT
 
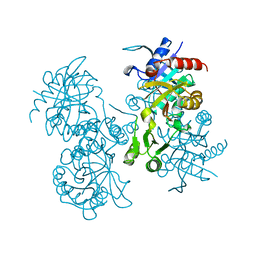 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | 分子名称: | GLUTATHIONE BIOSYNTHETIC LIGASE | | 著者 | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | 登録日 | 1995-05-16 | | 公開日 | 1995-07-31 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
5HXI
 
 | |
3VZQ
 
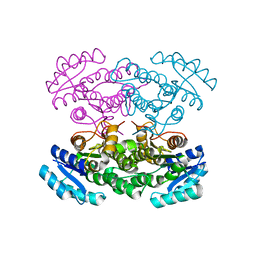 | |
1TMN
 
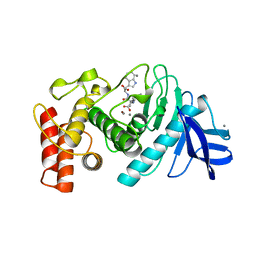 | |
3VZR
 
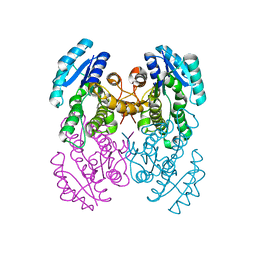 | |
3VND
 
 | | Crystal structure of tryptophan synthase alpha-subunit from the psychrophile Shewanella frigidimarina K14-2 | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, Tryptophan synthase alpha chain | | 著者 | Mitsuya, D, Tanaka, S, Matsumura, H, Takano, K, Urano, N, Ishida, M. | | 登録日 | 2012-01-12 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Strategy for cold adaptation of the tryptophan synthase alpha subunit from the psychrophile Shewanella frigidimarina K14-2: crystal structure and physicochemical properties
J.Biochem., 155, 2014
|
|
1TLP
 
 | | CRYSTALLOGRAPHIC STRUCTURAL ANALYSIS OF PHOSPHORAMIDATES AS INHIBITORS AND TRANSITION-STATE ANALOGS OF THERMOLYSIN | | 分子名称: | CALCIUM ION, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, THERMOLYSIN, ... | | 著者 | Tronrud, D.E, Monzingo, A.F, Matthews, B.W. | | 登録日 | 1987-06-29 | | 公開日 | 1989-01-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin.
Eur.J.Biochem., 157, 1986
|
|
3VZP
 
 | | Crystal structure of PhaB from Ralstonia eutropha | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, Acetoacetyl-CoA reductase, GLYCEROL, ... | | 著者 | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | 登録日 | 2012-10-15 | | 公開日 | 2013-08-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
3VZS
 
 | | Crystal structure of PhaB from Ralstonia eutropha in complex with Acetoacetyl-CoA and NADP | | 分子名称: | ACETOACETYL-COENZYME A, Acetoacetyl-CoA reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | 登録日 | 2012-10-15 | | 公開日 | 2013-08-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
3A56
 
 | |
3A54
 
 | |
1UJP
 
 | | Crystal Structure of Tryptophan Synthase A-Subunit From Thermus thermophilus HB8 | | 分子名称: | CITRIC ACID, Tryptophan synthase alpha chain | | 著者 | Asada, Y, Yokoyama, S, Kuramitsu, S, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-08-08 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Stabilization mechanism of the tryptophan synthase alpha-subunit from Thermus thermophilus HB8: X-ray crystallographic analysis and calorimetry.
J.Biochem.(Tokyo), 138, 2005
|
|
3VUQ
 
 | | Crystal structure of TTHA0167, a transcriptional regulator, TetR/AcrR family from Thermus thermophilus HB8 | | 分子名称: | Transcriptional regulator (TetR/AcrR family) | | 著者 | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | 登録日 | 2012-07-04 | | 公開日 | 2013-02-27 | | 最終更新日 | 2013-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure and function of a TetR family transcriptional regulator, SbtR, from thermus thermophilus HB8
Proteins, 81, 2013
|
|
3A55
 
 | |
2ZK9
 
 | | Crystal Structure of Protein-glutaminase | | 分子名称: | GLYCEROL, Protein-glutaminase, SODIUM ION | | 著者 | Hashizume, R. | | 登録日 | 2008-03-13 | | 公開日 | 2009-03-17 | | 最終更新日 | 2012-08-29 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Crystal structures of protein glutaminase and its pro forms converted into enzyme-substrate complex
J.Biol.Chem., 286, 2011
|
|
1OD6
 
 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | 分子名称: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | 著者 | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | 登録日 | 2003-02-13 | | 公開日 | 2003-03-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2D0W
 
 | | Crystal structure of cytochrome cL from Hyphomicrobium denitrificans | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, cytochrome cL | | 著者 | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | 登録日 | 2005-08-10 | | 公開日 | 2006-08-10 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
2D0V
 
 | | Crystal structure of methanol dehydrogenase from Hyphomicrobium denitrificans | | 分子名称: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, methanol dehydrogenase large subunit, ... | | 著者 | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | 登録日 | 2005-08-09 | | 公開日 | 2006-08-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
7JMA
 
 | |
7JMB
 
 | | Crystal structure of Nitrogenase iron-molybdenum cofactor biosynthesis enzyme NifB from Methanothermobacter thermautotrophicus with three Fe4S4 clusters | | 分子名称: | IRON/SULFUR CLUSTER, Nitrogenase iron-molybdenum cofactor biosynthesis protein NifB | | 著者 | Kang, W, Rettberg, L, Ribbe, M.W, Hu, Y. | | 登録日 | 2020-07-31 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-Ray Crystallographic Analysis of NifB with a Full Complement of Clusters: Structural Insights into the Radical SAM-Dependent Carbide Insertion During Nitrogenase Cofactor Assembly.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
