8PGL
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3027 | | 分子名称: | 7-[(1~{S})-1-[5-(carbamimidamidomethyl)-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 3027
To Be Published
|
|
8PGQ
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3225 | | 分子名称: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-[3-cyano-4-(methylsulfonylamino)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 3225
To Be Published
|
|
8PTV
 
 | |
8PH1
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3708 | | 分子名称: | 7-[(1~{S})-1-[(5~{R})-5-[(2~{R})-3-azanyl-2-methyl-propyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-(1~{H}-pyrazol-4-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 3708
To Be Published
|
|
8PGI
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2984 | | 分子名称: | 7-[(1~{S})-1-[5-[(3-azanyl-3-methyl-azetidin-1-yl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 2984
To Be Published
|
|
8PH0
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3637 | | 分子名称: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(2-oxidanylidene-1,3-dihydroindol-5-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 3637
To Be Published
|
|
8PGJ
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2985 | | 分子名称: | 7-[(1~{S})-1-[5-[(3-azanylazetidin-1-yl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 2985
To Be Published
|
|
8PG5
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2731 | | 分子名称: | 3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-7-[(1~{S})-1-(piperidin-4-ylmethoxy)ethyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 2731
To Be Published
|
|
8PG7
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2755 | | 分子名称: | 7-[(1~{S})-1-[3-(4-azanylbutyl)-2,5-bis(oxidanylidene)imidazolidin-1-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 2755
To Be Published
|
|
8PGW
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3460 | | 分子名称: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(5-methoxy-6-oxidanyl-pyridin-3-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 3460
To Be Published
|
|
8PY5
 
 | |
8PY9
 
 | | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 3s O2 exposure | | 分子名称: | (2S)-2-azanyl-6-[[(2R)-1-[[(2R)-3-methyl-1-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-3-sulfanylidene-propan-2-yl]amino]-6-oxidanylidene-hexanoic acid, FE (III) ION, Isopenicillin N synthase, ... | | 著者 | Rabe, P, Schofield, C.J. | | 登録日 | 2023-07-25 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 3s O2 exposure
To Be Published
|
|
8PY7
 
 | |
8PY6
 
 | |
8PXZ
 
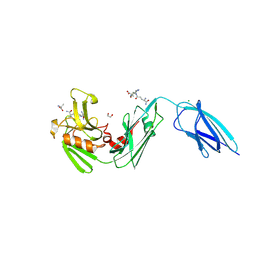 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with natural substrate | | 分子名称: | (2~{R})-2-[[(2~{S},6~{R})-6,7-bis(azanyl)-2-[[(4~{R})-4,5-bis(azanyl)-5-oxidanylidene-pentanoyl]amino]-7-oxidanylidene-heptanoyl]amino]propanoic acid, (2~{R})-2-azanyl-~{N}'-[(2~{S},6~{R})-6,7-bis(azanyl)-1-oxidanyl-7-oxidanylidene-heptan-2-yl]pentanediamide, 1,2-ETHANEDIOL, ... | | 著者 | de Munnik, M, Schofield, C.J. | | 登録日 | 2023-07-24 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with natural substrate
To Be Published
|
|
8PY8
 
 | |
8PXY
 
 | |
8PYA
 
 | | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 4s O2 exposure | | 分子名称: | (2S)-2-azanyl-6-[[(2R)-1-[[(2R)-3-methyl-1-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-3-sulfanylidene-propan-2-yl]amino]-6-oxidanylidene-hexanoic acid, FE (III) ION, Isopenicillin N synthase, ... | | 著者 | Rabe, P, Schofield, C.J. | | 登録日 | 2023-07-25 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 4s O2 exposure
To Be Published
|
|
8P7P
 
 | |
8P48
 
 | |
7E8Z
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with SS81 | | 分子名称: | 2-[[6-[(4-nitrophenyl)amino]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | 著者 | Tam, N.Y, Ng, Y.M, Shishodia, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | 登録日 | 2021-03-03 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7DYV
 
 | | Human JMJD5 in complex with MN and 5-(benzylamino)pyridine-2,4-dicarboxylic acid. | | 分子名称: | 5-(benzylamino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | 著者 | Nakashima, Y, Brewitz, L, Schofield, C.J. | | 登録日 | 2021-01-23 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYW
 
 | | Human JMJD5 in complex with MN and 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | 分子名称: | 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | 著者 | Nakashima, Y, Brewitz, L, Schofield, C.J. | | 登録日 | 2021-01-23 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYT
 
 | | Human JMJD5 in complex with MN and 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | 分子名称: | 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | 著者 | Nakashima, Y, Brewitz, L, Schofield, C.J. | | 登録日 | 2021-01-23 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYX
 
 | | Human JMJD5 in complex with MN and 5-((2-cyclopropylbenzyl)amino)pyridine-2,4-dicarboxylic acid. | | 分子名称: | 5-((2-cyclopropylbenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | 著者 | Nakashima, Y, Brewitz, L, Schofield, C.J. | | 登録日 | 2021-01-23 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
