4RM0
 
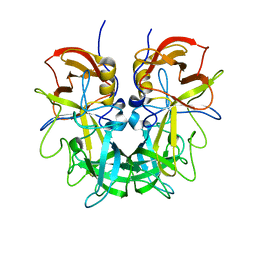 | | Crystal structure of Norovirus OIF P domain in complex with Lewis a trisaccharide | | 分子名称: | Capsid protein, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Liu, W, Chen, Y, Tan, M, Xia, M, Li, X, Jiang, X, Rao, Z. | | 登録日 | 2014-10-18 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface.
Plos Pathog., 11, 2015
|
|
4RSU
 
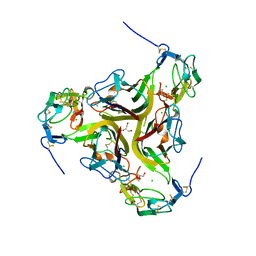 | | Crystal structure of the light and hvem complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Liu, W, Ramagoal, U.A, Himmel, D, Bonanno, J.B, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-11-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
4RLZ
 
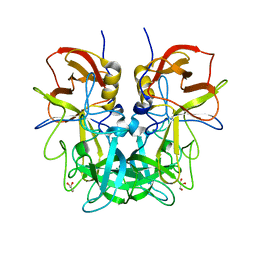 | | Crystal structure of Norovirus OIF P domain | | 分子名称: | Capsid protein, GLYCEROL | | 著者 | Liu, W, Chen, Y, Tan, M, Xia, M, Li, X, Jiang, X, Rao, Z. | | 登録日 | 2014-10-18 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface.
Plos Pathog., 11, 2015
|
|
1SF2
 
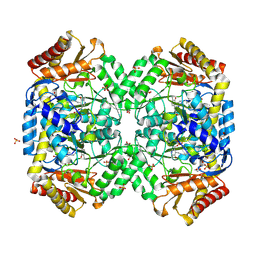 | | Structure of E. coli gamma-aminobutyrate aminotransferase | | 分子名称: | 1,2-ETHANEDIOL, 4-aminobutyrate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | 登録日 | 2004-02-19 | | 公開日 | 2004-09-14 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
1SZS
 
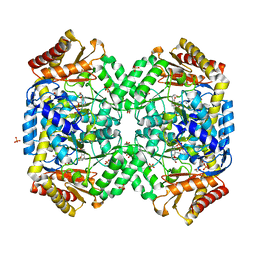 | | The structure of gamma-aminobutyrate aminotransferase mutant: I50Q | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | 著者 | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | 登録日 | 2004-04-06 | | 公開日 | 2005-03-01 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1SZU
 
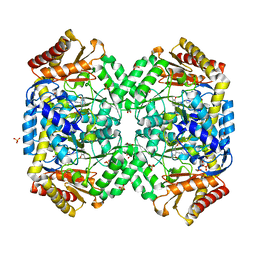 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | 著者 | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | 登録日 | 2004-04-06 | | 公開日 | 2005-03-01 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
3HXI
 
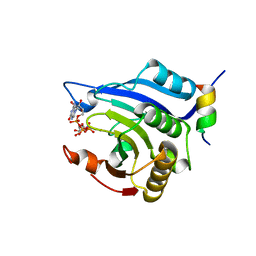 | | Crystal structure of Schistosome eIF4E complexed with m7GpppG and 4E-BP | | 分子名称: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Eukaryotic Translation Initiation 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | 著者 | Liu, W, Zhao, R, Jones, D.N.M, Davis, R.E. | | 登録日 | 2009-06-20 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into parasite EIF4E binding specificity for m7G and m2,2,7G mRNA cap.
J.Biol.Chem., 284, 2009
|
|
1XD5
 
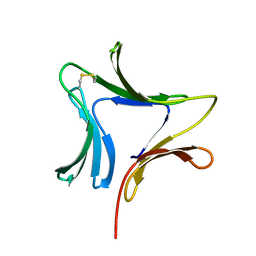 | | Crystal structures of novel monomeric monocot mannose-binding lectins from Gastrodia elata | | 分子名称: | SULFATE ION, antifungal protein GAFP-1 | | 著者 | Liu, W, Yang, N, Wang, M, Huang, R.H, Hu, Z, Wang, D.C. | | 登録日 | 2004-09-04 | | 公開日 | 2005-01-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Mechanism Governing the Quaternary Organization of Monocot Mannose-binding Lectin Revealed by the Novel Monomeric Structure of an Orchid Lectin
J.Biol.Chem., 280, 2005
|
|
1XD6
 
 | | Crystal structures of novel monomeric monocot mannose-binding lectins from Gastrodia elata | | 分子名称: | SULFATE ION, gastrodianin-4 | | 著者 | Liu, W, Yang, N, Wang, M, Huang, R.H, Hu, Z, Wang, D.C. | | 登録日 | 2004-09-04 | | 公開日 | 2005-01-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Mechanism Governing the Quaternary Organization of Monocot Mannose-binding Lectin Revealed by the Novel Monomeric Structure of an Orchid Lectin
J.Biol.Chem., 280, 2005
|
|
5L19
 
 | |
3HXG
 
 | | Crystal structure of Schistsome eIF4E complexed with m7GpppA and 4E-BP | | 分子名称: | Eukaryotic Translation Initiation Factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | 著者 | Liu, W, Zhao, R, Jones, D.N.M, Davis, R.E. | | 登録日 | 2009-06-20 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into parasite EIF4E binding specificity for m7G and m2,2,7G mRNA cap.
J.Biol.Chem., 284, 2009
|
|
1ZOB
 
 | |
5L36
 
 | |
1ZOD
 
 | |
4DX9
 
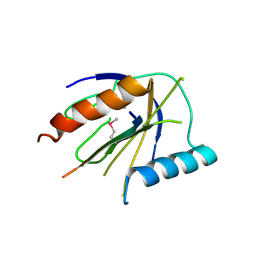 | | ICAP1 in complex with integrin beta 1 cytoplasmic tail | | 分子名称: | Integrin beta-1, Integrin beta-1-binding protein 1 | | 著者 | Liu, W, Draheim, K, Zhang, R, Calderwood, D.A, Boggon, T.J. | | 登録日 | 2012-02-27 | | 公開日 | 2013-01-09 | | 最終更新日 | 2020-09-02 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation.
Mol.Cell, 49, 2013
|
|
4EIY
 
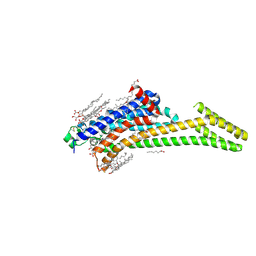 | | Crystal structure of the chimeric protein of A2aAR-BRIL in complex with ZM241385 at 1.8A resolution | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | 著者 | Liu, W, Chun, E, Thompson, A.A, Chubukov, P, Xu, F, Katritch, V, Han, G.W, Heitman, L.H, Ijzerman, A.P, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | 登録日 | 2012-04-06 | | 公開日 | 2012-07-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for allosteric regulation of GPCRs by sodium ions.
Science, 337, 2012
|
|
4FHQ
 
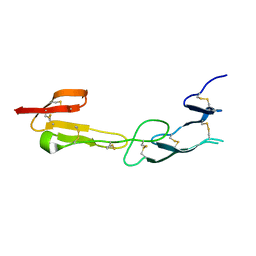 | | Crystal Structure of HVEM | | 分子名称: | Tumor necrosis factor receptor superfamily member 14 | | 著者 | Liu, W, Zhan, C, Patskovsky, Y, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-06-06 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Increased Heterologous Protein Expression in Drosophila S2 Cells for Massive Production of Immune Ligands/Receptors and Structural Analysis of Human HVEM.
Mol Biotechnol, 57, 2015
|
|
2HGZ
 
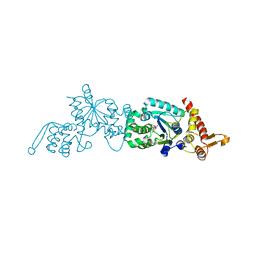 | | Crystal structure of a p-benzoyl-L-phenylalanyl-tRNA synthetase | | 分子名称: | PARA-(BENZOYL)-PHENYLALANINE, Tyrosyl-tRNA synthetase | | 著者 | Liu, W, Alfonta, L, Mack, A.V, Schultz, P.G. | | 登録日 | 2006-06-27 | | 公開日 | 2007-06-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the recognition of para-benzoyl-L-phenylalanine by evolved aminoacyl-tRNA synthetases.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2P4W
 
 | | Crystal structure of heat shock regulator from Pyrococcus furiosus | | 分子名称: | SULFATE ION, Transcriptional regulatory protein arsR family | | 著者 | Liu, W, Vierke, G, Panjikar, S, Thomm, M, Ladenstein, R. | | 登録日 | 2007-03-13 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of the Archaeal Heat Shock Regulator from Pyrococcus furiosus: A Molecular Chimera Representing Eukaryal and Bacterial Features.
J.Mol.Biol., 369, 2007
|
|
6NGG
 
 | |
6NG3
 
 | | Crystal structure of human CD160 and HVEM complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen,Tumor necrosis factor receptor superfamily member 14, MAGNESIUM ION, ... | | 著者 | Liu, W, Bonanno, J, Almo, S.C. | | 登録日 | 2018-12-21 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Structural Basis of CD160:HVEM Recognition.
Structure, 27, 2019
|
|
6NG9
 
 | |
3ULR
 
 | | Lysozyme contamination facilitates crystallization of a hetero-trimericCortactin:Arg:Lysozyme complex | | 分子名称: | Abelson tyrosine-protein kinase 2, Lysozyme C, Src substrate cortactin | | 著者 | Liu, W, MacGrath, S, Koleske, A.J, Boggon, T.J. | | 登録日 | 2011-11-11 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Lysozyme contamination facilitates crystallization of a heterotrimeric cortactin-Arg-lysozyme complex.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6AE4
 
 | |
6AE6
 
 | |
