7W73
 
 | | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | 登録日 | 2021-12-03 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6T
 
 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | 登録日 | 2022-06-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6U
 
 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | 登録日 | 2022-06-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6V
 
 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | 登録日 | 2022-06-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6S
 
 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | 登録日 | 2022-06-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7WCC
 
 | | Oxidase ChaP-D49L/Q91C mutant | | 分子名称: | ChaP, FE (III) ION | | 著者 | Sun, M.X, Zheng, W, Wang, Y.S, Zhu, J.P, Tan, R.X. | | 登録日 | 2021-12-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Oxidase ChaP-D49L/Q91C mutant
To Be Published
|
|
3KN0
 
 | | Structure of BACE bound to SCH708236 | | 分子名称: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3KMX
 
 | | Structure of BACE bound to SCH346572 | | 分子名称: | 4-butoxy-3-chlorobenzyl imidothiocarbamate, Beta-secretase 1 | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3KMY
 
 | | Structure of BACE bound to SCH12472 | | 分子名称: | 3-[2-(3-chlorophenyl)ethyl]pyridin-2-amine, Beta-secretase 1 | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
6A52
 
 | | Oxidase ChaP-H1 | | 分子名称: | FE (II) ION, dioxidase ChaP-H1 | | 著者 | Zhang, B, Ge, H.M. | | 登録日 | 2018-06-21 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A4Z
 
 | | Oxidase ChaP | | 分子名称: | ChaP protein, FE (II) ION | | 著者 | Zhang, B, Ge, H.M. | | 登録日 | 2018-06-21 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
7WB2
 
 | | Oxidase ChaP-D49L/Y109F mutant | | 分子名称: | ChaP, FE (III) ION | | 著者 | Zong, Y, Zheng, W, Wang, Y, Zhu, J, Tan, R. | | 登録日 | 2021-12-15 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7W5E
 
 | | Oxidase ChaP D49L mutant | | 分子名称: | ChaP, FE (III) ION | | 著者 | Wang, Y, Zheng, W, Meng, Z, Jin, Y, Zhu, J, Tan, R. | | 登録日 | 2021-11-30 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6VW1
 
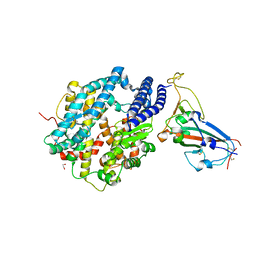 | | Structure of SARS-CoV-2 chimeric receptor-binding domain complexed with its receptor human ACE2 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Shang, J, Ye, G, Shi, K, Wan, Y.S, Aihara, H, Li, F. | | 登録日 | 2020-02-18 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural basis of receptor recognition by SARS-CoV-2.
Nature, 581, 2020
|
|
6VSJ
 
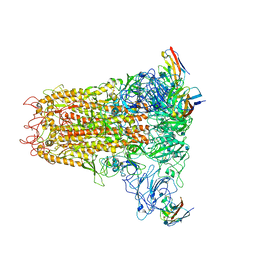 | | Cryo-electron microscopy structure of mouse coronavirus spike protein complexed with its murine receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 1, Spike glycoprotein | | 著者 | Shang, J, Wan, Y.S, Liu, C, Yount, B, Gully, K, Yang, Y, Auerbach, A, Peng, G.Q, Baric, R, Li, F. | | 登録日 | 2020-02-11 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.94 Å) | | 主引用文献 | Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry.
Plos Pathog., 16, 2020
|
|
7YMV
 
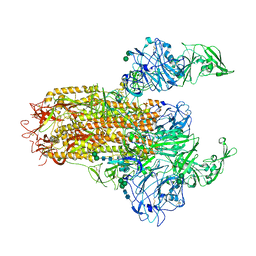 | | Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | 登録日 | 2022-07-29 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (6.74 Å) | | 主引用文献 | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMW
 
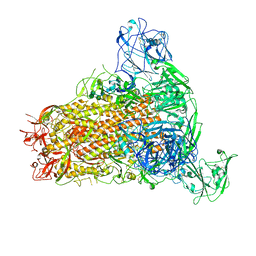 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | 登録日 | 2022-07-29 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (6.05 Å) | | 主引用文献 | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMX
 
 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | 登録日 | 2022-07-29 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.44 Å) | | 主引用文献 | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMT
 
 | | Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | 登録日 | 2022-07-29 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (6.55 Å) | | 主引用文献 | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YN0
 
 | | Cryo-EM structure of MERS-CoV spike protein, all RBD-down conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | 登録日 | 2022-07-29 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMY
 
 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | 登録日 | 2022-07-29 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.96 Å) | | 主引用文献 | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMZ
 
 | | Cryo-EM structure of MERS-CoV spike protein, intermediate conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | 登録日 | 2022-07-29 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (4.39 Å) | | 主引用文献 | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
1FHS
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE SRC HOMOLOGY DOMAIN-2 OF THE GROWTH FACTOR RECEPTOR BOUND PROTEIN-2, NMR, 18 STRUCTURES | | 分子名称: | GROWTH FACTOR RECEPTOR BOUND PROTEIN-2 | | 著者 | Senior, M.M, Frederick, A.F, Black, S, Perkins, L.M, Wilson, O, Snow, M.E, Wang, Y.-S. | | 登録日 | 1997-06-12 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional solution structure of the Src homology domain-2 of the growth factor receptor-bound protein-2.
J.Biomol.NMR, 11, 1998
|
|
3L5B
 
 | | Structure of BACE Bound to SCH713601 | | 分子名称: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L59
 
 | | Structure of BACE Bound to SCH710413 | | 分子名称: | (2Z)-3-(3-chlorobenzyl)-2-imino-5,5-dimethylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
