6KNS
 
 | | Crystal structure of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis (space group I4122) | | 分子名称: | CALCIUM ION, Putative metal-dependent hydrolase, ZINC ION | | 著者 | Na, H.W, Namgung, B, Song, W.S, Yoon, S.I. | | 登録日 | 2019-08-07 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural and biochemical analyses of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
1SO6
 
 | | Crystal structure of E112Q/H136A double mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | 分子名称: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | 著者 | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | 登録日 | 2004-03-12 | | 公開日 | 2004-06-08 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SO5
 
 | | Crystal structure of E112Q mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | 分子名称: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | 著者 | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | 登録日 | 2004-03-12 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
6KNT
 
 | | Crystal structure of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis (space group P4332) | | 分子名称: | Putative metal-dependent hydrolase, ZINC ION | | 著者 | Na, H.W, Namgung, B, Song, W.S, Yoon, S.I. | | 登録日 | 2019-08-07 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and biochemical analyses of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
1SO3
 
 | | Crystal structure of H136A mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | 分子名称: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | 著者 | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | 登録日 | 2004-03-12 | | 公開日 | 2004-06-08 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
6WYA
 
 | | RTX (Reverse Transcription Xenopolymerase) in complex with a DNA duplex and dAMPNPP | | 分子名称: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | 登録日 | 2020-05-12 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1SO4
 
 | | Crystal structure of K64A mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | 分子名称: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | 著者 | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | 登録日 | 2004-03-12 | | 公開日 | 2004-06-08 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
6JV6
 
 | | Crystal structure of the sirohydrochlorin chelatase SirB from Bacillus subtilis subspecies spizizenii in complex with cobalt | | 分子名称: | COBALT (II) ION, Sirohydrochlorin ferrochelatase | | 著者 | Nam, M.S, Song, W.S, Park, S.C, Yoon, S.I. | | 登録日 | 2019-04-16 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Cobalt complex structure of the sirohydrochlorin chelatase SirB from Bacillus subtilis subsp. spizizenii.
KOREAN J MICROBIOL., 55, 2019
|
|
6WYB
 
 | | RTX (Reverse Transcription Xenopolymerase) in complex with an RNA/DNA hybrid | | 分子名称: | 1,2-ETHANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, DNA polymerase, ... | | 著者 | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | 登録日 | 2020-05-12 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C7Z
 
 | |
6JSS
 
 | |
6JYI
 
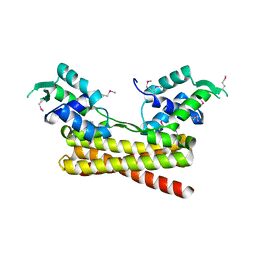 | | Crystal structure of the PadR-like transcriptional regulator BC1756 from Bacillus cereus | | 分子名称: | Transcriptional repressor PadR | | 著者 | Kim, T.H, Park, S.C, Lee, K.C, Song, W.S, Yoon, S.I. | | 登録日 | 2019-04-26 | | 公開日 | 2019-06-26 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural and DNA-binding studies of the PadR-like transcriptional regulator BC1756 from Bacillus cereus.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
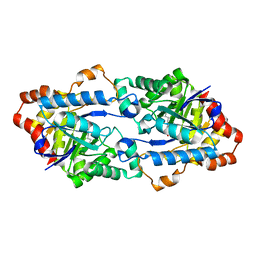
6JSU
 
 | |
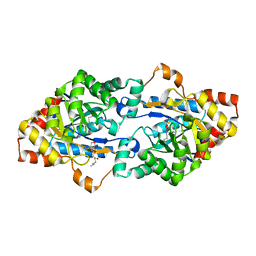
6JST
 
 | |
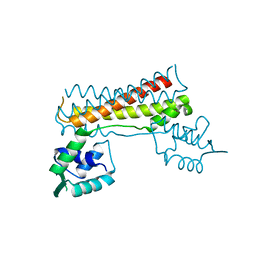
7CBV
 
 | |
7DHQ
 
 | | Structure of Halothiobacillus neapolitanus Microcompartments Protein CsoS1D | | 分子名称: | Microcompartments protein | | 著者 | Xue, B, Tan, Y.Q, Ali, S, Robinson, R.C, Narita, A, Yew, W.S. | | 登録日 | 2020-11-17 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
7CKB
 
 | | Simplified Alpha-Carboxysome, T=3 | | 分子名称: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | 著者 | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | 登録日 | 2020-07-16 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
7CKC
 
 | | Simplified Alpha-Carboxysome, T=4 | | 分子名称: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | 著者 | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | 登録日 | 2020-07-16 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
7DWM
 
 | | Crystal structure of the phage VqmA-DPO complex | | 分子名称: | 3,5-dimethylpyrazin-2-ol, Transcriptional regulator | | 著者 | Gu, Y, Yang, W.S. | | 登録日 | 2021-01-17 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Understanding the mechanism of asymmetric gene regulation determined by the VqmA of vibriophage.
Biochem.Biophys.Res.Commun., 558, 2021
|
|
7E8Z
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with SS81 | | 分子名称: | 2-[[6-[(4-nitrophenyl)amino]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | 著者 | Tam, N.Y, Ng, Y.M, Shishodia, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | 登録日 | 2021-03-03 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
1WVE
 
 | | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon its Binding to the Cytochrome Subunit | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-cresol dehydrogenase [hydroxylating] cytochrome c subunit, 4-cresol dehydrogenase [hydroxylating] flavoprotein subunit, ... | | 著者 | Cunane, L.M, Chen, Z.-W, McIntire, W.S, Mathews, F.S. | | 登録日 | 2004-12-15 | | 公開日 | 2005-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon Its Binding to the Cytochrome Subunit
Biochemistry, 44, 2005
|
|
1WVF
 
 | | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon its Binding to the Cytochrome Subunit | | 分子名称: | 4-cresol dehydrogenase [hydroxylating] flavoprotein subunit, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Cunane, L.M, Chen, Z.-W, McIntire, W.S, Mathews, F.S. | | 登録日 | 2004-12-15 | | 公開日 | 2005-03-08 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon Its Binding to the Cytochrome Subunit
Biochemistry, 44, 2005
|
|
1CMA
 
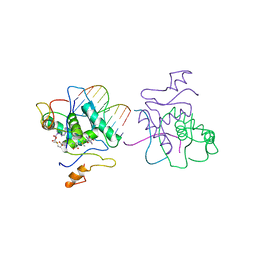 | | MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE | | 分子名称: | DNA (5'-D(*AP*GP*AP*CP*GP*TP*CP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*AP*CP*GP*TP*CP*T)-3'), PROTEIN (MET REPRESSOR), ... | | 著者 | Somers, W.S, Phillips, S.E.V. | | 登録日 | 1992-08-24 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands.
Nature, 359, 1992
|
|
1DII
 
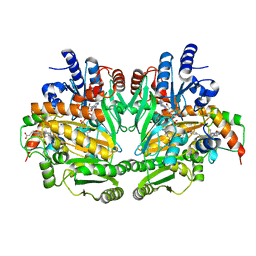 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE AT 2.5 A RESOLUTION | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | 著者 | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.N, McIntire, W.S. | | 登録日 | 1999-11-29 | | 公開日 | 1999-12-08 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1DIQ
 
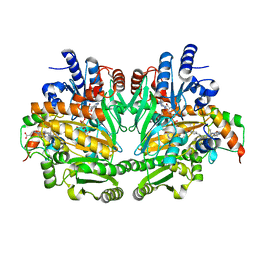 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE WITH SUBSTRATE BOUND | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | 著者 | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.S, McIntire, W.S. | | 登録日 | 1999-11-29 | | 公開日 | 1999-12-08 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
