3F6K
 
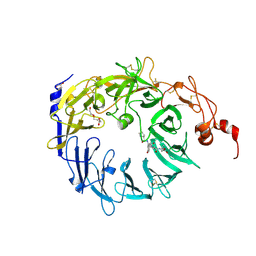 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with neurotensin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Neurotensin, ... | | 著者 | Quistgaard, E.M, Madsen, P, Groftehauge, M.K, Nissen, P, Petersen, C.M, Thirup, S. | | 登録日 | 2008-11-06 | | 公開日 | 2008-12-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Ligands bind to Sortilin in the tunnel of a ten-bladed beta-propeller domain.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4UZ3
 
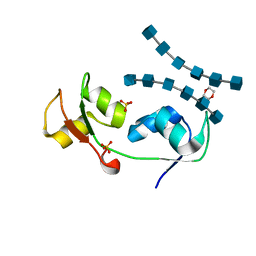 | | Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | 著者 | Wong, J.E.M.M, Blaise, M. | | 登録日 | 2014-09-04 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | An Intermolecular Binding Mechanism Involving Multiple Lysm Domains Mediates Carbohydrate Recognition by an Endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UZ2
 
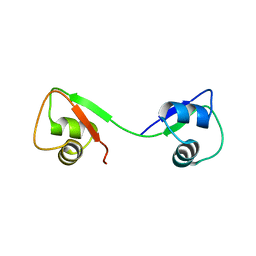 | |
1OB2
 
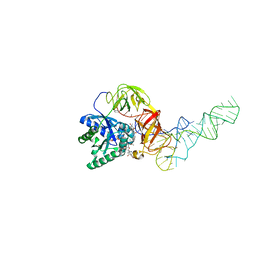 | | E. coli elongation factor EF-Tu complexed with the antibiotic kirromycin, a GTP analog, and Phe-tRNA | | 分子名称: | ELONGATION FACTOR TU, KIRROMYCIN, MAGNESIUM ION, ... | | 著者 | Kristensen, O, Nissen, P, Nyborg, J. | | 登録日 | 2003-01-24 | | 公開日 | 2004-05-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Kirromycin Defines a Specific Domain Arrangement of Elongation Factor EF-TU
To be Published
|
|
1ZC8
 
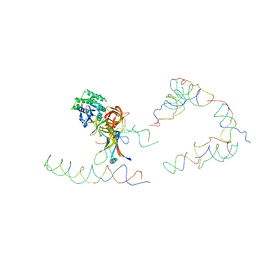 | | Coordinates of tmRNA, SmpB, EF-Tu and h44 fitted into Cryo-EM map of the 70S ribosome and tmRNA complex | | 分子名称: | Elongation factor Tu, H2 16S rRNA, H2b d mRNA, ... | | 著者 | Valle, M, Gillet, R, Kaur, S, Henne, A, Ramakrishnan, V, Frank, J. | | 登録日 | 2005-04-11 | | 公開日 | 2005-04-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (13 Å) | | 主引用文献 | Visualizing tmRNA Entry into a Stalled Ribosome
Science, 300, 2003
|
|
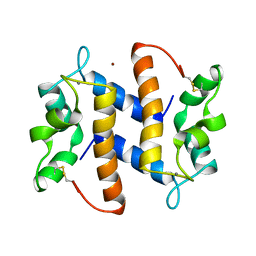
3PSR
 
 | |
2PSR
 
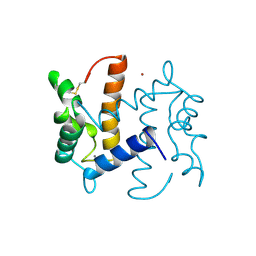 | |
1PSR
 
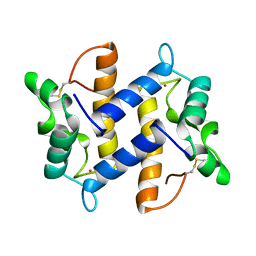 | | HUMAN PSORIASIN (S100A7) | | 分子名称: | HOLMIUM ATOM, PSORIASIN | | 著者 | Brodersen, D.E, Etzerodt, M, Madsen, P, Celis, J, Thoegersen, H.C, Nyborg, J, Kjeldgaard, M. | | 登録日 | 1997-11-27 | | 公開日 | 1999-01-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | EF-hands at atomic resolution: the structure of human psoriasin (S100A7) solved by MAD phasing.
Structure, 6, 1998
|
|
