9B3P
 
 | |
5HOC
 
 | | p73 homo-tetramerization domain mutant II | | 分子名称: | Tumor protein p73 | | 著者 | Coutandin, D, Krojer, T, Salah, E, Mathea, S, Sumyk, M, Knapp, S, Dotsch, V. | | 登録日 | 2016-01-19 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.36007786 Å) | | 主引用文献 | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
7M1X
 
 | |
4QOZ
 
 | |
6KYB
 
 | | Crystal structure of Atg18 from Saccharomyces cerevisiae | | 分子名称: | Autophagy-related protein 18 | | 著者 | Tang, D, Lei, Y, Liao, G, Chen, Q, Xu, L, Lu, K, Qi, S. | | 登録日 | 2019-09-17 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of Atg18 reveals a new binding site for Atg2 in Saccharomyces cerevisiae.
Cell.Mol.Life Sci., 78, 2021
|
|
6SQG
 
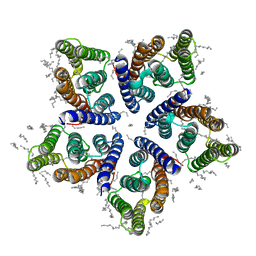 | | Crystal structure of viral rhodopsin OLPVRII | | 分子名称: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | 著者 | Gushchin, I, Kovalev, K, Bratanov, D, Polovinkin, V, Astashkin, R, Popov, A, Bourenkov, G, Gordeliy, V. | | 登録日 | 2019-09-03 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unique structure and function of viral rhodopsins.
Nat Commun, 10, 2019
|
|
7F3Y
 
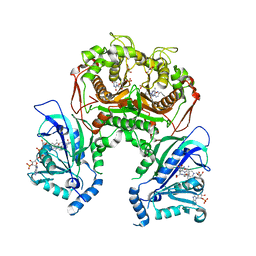 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with methotrexate (MTX), NADPH and dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | 著者 | Vanichtanankul, J, Tanramluk, D, Yuvaniyama, J, Yuthavong, Y. | | 登録日 | 2021-06-17 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.252 Å) | | 主引用文献 | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
7F3Z
 
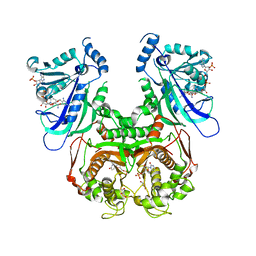 | | Double mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS-K1, C59R+S108N) complexed with Trimethoprim (TOP), NADPH and dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | 著者 | Vanichtanankul, J, Tanramluk, D, Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | 登録日 | 2021-06-17 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
5HOB
 
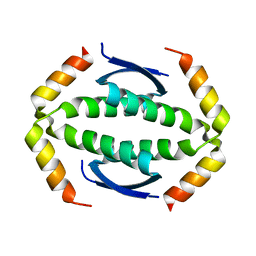 | | p73 homo-tetramerization domain mutant I | | 分子名称: | MAGNESIUM ION, Tumor protein p73 | | 著者 | Coutandin, D, Krojer, T, Salah, E, Mathea, S, Knapp, S, Dotsch, V. | | 登録日 | 2016-01-19 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.220013 Å) | | 主引用文献 | Structural basis of p63/p73 hetero-tetramerization
To Be Published
|
|
4U01
 
 | | HCV NS3/4A serine protease in complex with 6570 | | 分子名称: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | 著者 | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | 登録日 | 2014-07-11 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6P7W
 
 | | Structure of the K. lactis CBF3 core - Ndc10 D1 complex | | 分子名称: | Cep3, Ctf13, Ndc10, ... | | 著者 | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | 登録日 | 2019-06-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6P7V
 
 | | Structure of the K. lactis CBF3 core | | 分子名称: | Cep3, Ctf13, Skp1 | | 著者 | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | 登録日 | 2019-06-06 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6P7X
 
 | | Structure of the K. lactis CBF3 core - Ndc10 D1D2 complex | | 分子名称: | Cep3, Ctf13, Ndc10, ... | | 著者 | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | 登録日 | 2019-06-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
3JCT
 
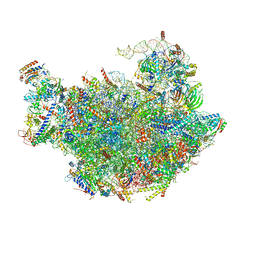 | | Cryo-em structure of eukaryotic pre-60S ribosomal subunits | | 分子名称: | 60S ribosomal protein L11-A, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | 著者 | Wu, S, Kumcuoglu, B, Yan, K.G, Brown, H, Zhang, Y.X, Tan, D, Gamalinda, M, Yuan, Y, Li, Z.F, Jakovljevic, J, Ma, C.Y, Lei, J.L, Dong, M.Q, Woolford Jr, J.L, Gao, N. | | 登録日 | 2016-03-09 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes
Nature, 534, 2016
|
|
3J2U
 
 | | Kinesin-13 KLP10A HD in complex with CS-tubulin and a microtubule | | 分子名称: | Kinesin-like protein Klp10A, Tubulin alpha-1A chain, Tubulin beta-2B chain | | 著者 | Asenjo, A.B, Chatterjee, C, Tan, D, DePaoli, V, Rice, W.J, Diaz-Avalos, R, Silvestry, M, Sosa, H. | | 登録日 | 2013-01-10 | | 公開日 | 2013-03-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (10.8 Å) | | 主引用文献 | Structural model for tubulin recognition and deformation by Kinesin-13 microtubule depolymerases.
Cell Rep, 3, 2013
|
|
9J1R
 
 | | Structure of a triple-helix region of human Collagen type II from Trautec | | 分子名称: | SULFATE ION, Triple-helix region of human collagen type II | | 著者 | Fan, X, Chu, Y, Zhai, Y, Fu, S, Li, D, Cao, K, Feng, P, Wang, X, Le, H, Tang, D, Zhang, F, Qian, S. | | 登録日 | 2024-08-05 | | 公開日 | 2024-08-21 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure of a triple-helix region of human Collagen type II from Trautec
To Be Published
|
|
8WIK
 
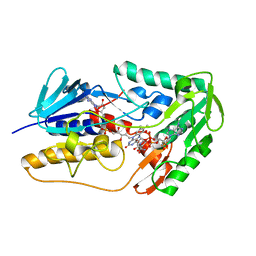 | | Crystal structure of human FSP1 | | 分子名称: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Feng, S, Huang, X, Tang, D, Qi, S. | | 登録日 | 2023-09-24 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of human ferroptosis suppressive protein 1 in complex with flavin adenine dinucleotide and nicotinamide adenine nucleotide.
MedComm (2020), 5, 2024
|
|
5G06
 
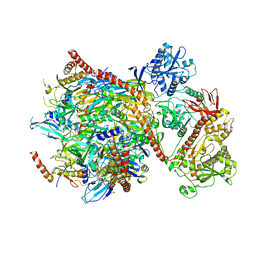 | | Cryo-EM structure of yeast cytoplasmic exosome | | 分子名称: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | 著者 | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | 登録日 | 2016-03-17 | | 公開日 | 2016-06-15 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
5H64
 
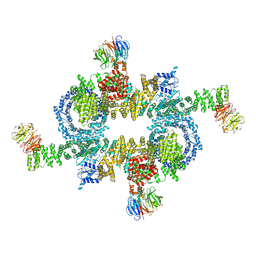 | | Cryo-EM structure of mTORC1 | | 分子名称: | Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex subunit LST8 | | 著者 | Yang, H, Wang, J, Liu, M, Chen, X, Huang, M, Tan, D, Dong, M, Wong, C.C.L, Wang, J, Xu, Y, Wang, H. | | 登録日 | 2016-11-10 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | 4.4 angstrom Resolution Cryo-EM structure of human mTOR Complex 1
Protein Cell, 7, 2016
|
|
6Z9M
 
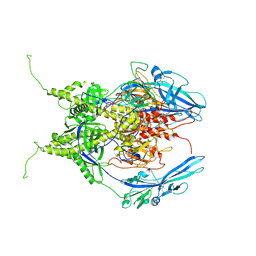 | | Pseudoatomic model of the pre-fusion conformation of glycoprotein B of Herpes simplex virus 1 | | 分子名称: | Envelope glycoprotein B | | 著者 | Vollmer, B, Prazak, V, Vasishtan, D, Jefferys, E.E, Hernandez-Duran, A, Vallbracht, M, Klupp, B, Mettenleiter, T.C, Backovic, M, Rey, F.A, Topf, M, Gruenewald, K. | | 登録日 | 2020-06-04 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (9.1 Å) | | 主引用文献 | The prefusion structure of herpes simplex virus glycoprotein B.
Sci Adv, 6, 2020
|
|
7VER
 
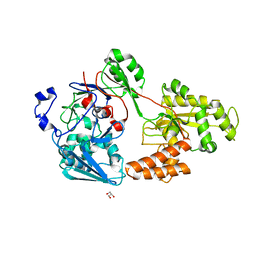 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a full open conformation | | 分子名称: | GLYCEROL, SPH1118 | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEU
 
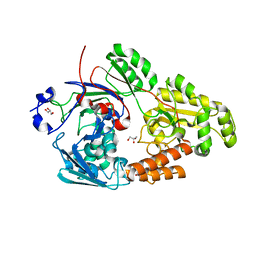 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with galacturonic acid | | 分子名称: | GLYCEROL, SPH1118, alpha-D-galactopyranuronic acid | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.736 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEW
 
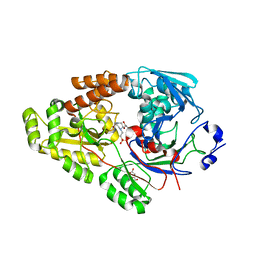 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with unsaturated trigalacturonic acid | | 分子名称: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEQ
 
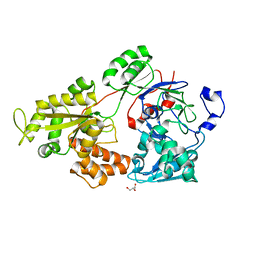 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in an open conformation | | 分子名称: | GLYCEROL, SPH1118 | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VET
 
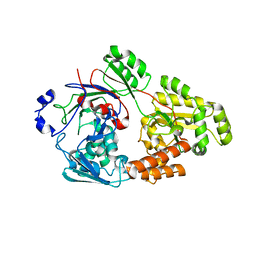 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a closed conformation | | 分子名称: | SPH1118 | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
