8FR6
 
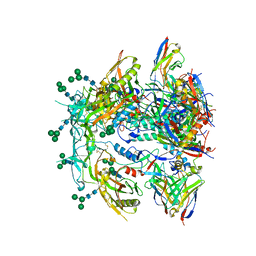 | | Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Wang, S, Kwong, P.D. | | 登録日 | 2023-01-06 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
7JWY
 
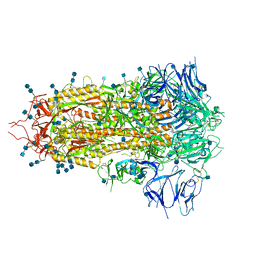 | | Structure of SARS-CoV-2 spike at pH 4.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Kwong, P.D. | | 登録日 | 2020-08-26 | | 公開日 | 2020-11-25 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6WNH
 
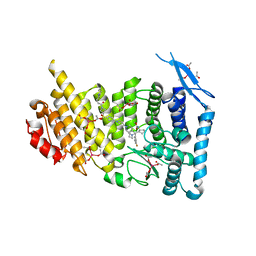 | | Menin bound to inhibitor M-808 | | 分子名称: | Menin, methyl [(1S,2R)-2-{(1S)-2-(azetidin-1-yl)-1-(3-fluorophenyl)-1-[1-({1-[4-({1-[4-(piperidin-1-yl)butanoyl]azetidin-3-yl}sulfonyl)phenyl]azetidin-3-yl}methyl)piperidin-4-yl]ethyl}cyclopentyl]carbamate, praseodymium triacetate | | 著者 | Stuckey, J.A. | | 登録日 | 2020-04-22 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of M-808 as a Highly Potent, Covalent, Small-Molecule Inhibitor of the Menin-MLL Interaction with StrongIn VivoAntitumor Activity.
J.Med.Chem., 63, 2020
|
|
6E1A
 
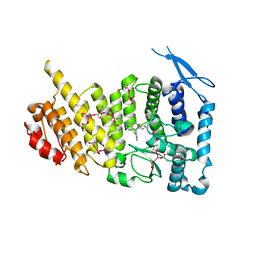 | | Menin bound to M-89 | | 分子名称: | (1S,2R)-2-[(4S)-2-methyl-4-{1-[(1-{4-[(pyridin-4-yl)sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}-1,2,3,4-tetrahydroisoquinolin-4-yl]cyclopentyl methylcarbamate, Menin, praseodymium triacetate | | 著者 | Stuckey, J.A. | | 登録日 | 2018-07-09 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure-Based Discovery of M-89 as a Highly Potent Inhibitor of the Menin-Mixed Lineage Leukemia (Menin-MLL) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
8G9X
 
 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Changela, A, Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.46 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8GAS
 
 | | vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-23 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.04 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G85
 
 | | vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-17 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9W
 
 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Changela, A, Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.66 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9Y
 
 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Changela, A, Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.28 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
3JWO
 
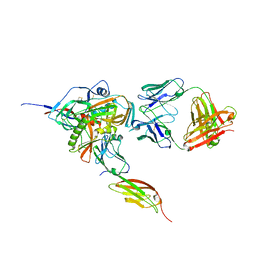 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D Heavy CHAIN, FAB 48D LIGHT CHAIN, ... | | 著者 | Pancera, M, Majeed, S, Huang, C.C, Kwon, Y.D, Zhou, T, Robinson, J.E, Sodroski, J, Wyatt, R, Kwong, P.D. | | 登録日 | 2009-09-18 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5ESV
 
 | | Crystal Structure of Broadly Neutralizing Antibody CH03, Isolated from Donor CH0219, in Complex with Scaffolded Trimeric HIV-1 Env V1V2 Domain from the Clade C Superinfecting Strain of Donor CAP256. | | 分子名称: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase,Envelope glycoprotein gp160, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gorman, J, Yang, M, Kwong, P.D. | | 登録日 | 2015-11-17 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.105 Å) | | 主引用文献 | Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5ESZ
 
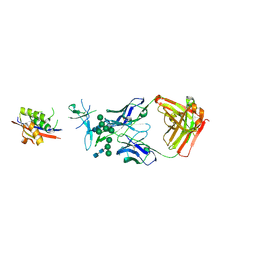 | | Crystal Structure of Broadly Neutralizing Antibody CH04, Isolated from Donor CH0219, in Complex with Scaffolded Trimeric HIV-1 Env V1V2 Domain from the Clade AE Strain A244 | | 分子名称: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase,Envelope glycoprotein gp160, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH04 Heavy Chain, ... | | 著者 | Gorman, J, Yang, M, Kwong, P.D. | | 登録日 | 2015-11-17 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (4.191 Å) | | 主引用文献 | Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design.
Nat. Struct. Mol. Biol., 23, 2016
|
|
6BWW
 
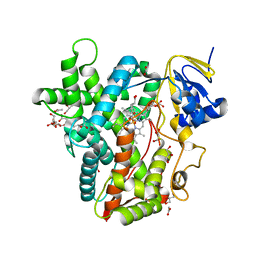 | |
7KNE
 
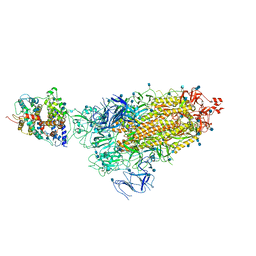 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNI
 
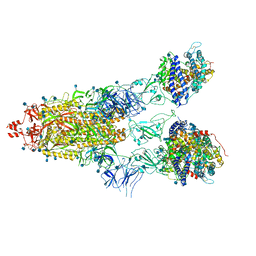 | | Cryo-EM structure of Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNB
 
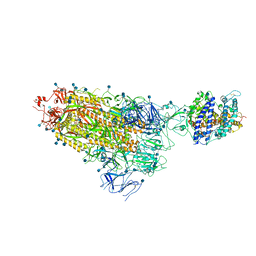 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMB
 
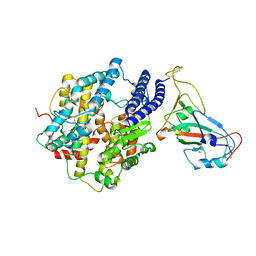 | | ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C3 for Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-02 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNH
 
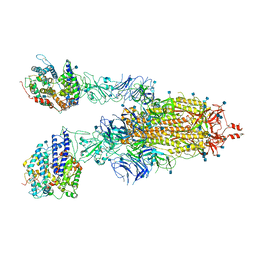 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMS
 
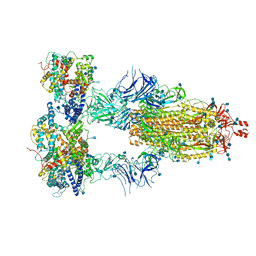 | | Cryo-EM structure of triple ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-03 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMZ
 
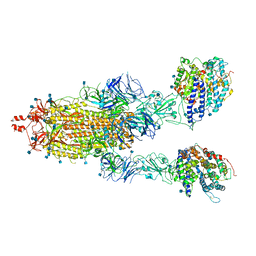 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-03 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
3LJM
 
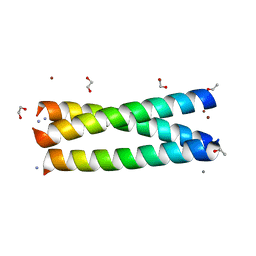 | | Structure of de novo designed apo peptide coil SER L9C | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, COIL SER L9C, ... | | 著者 | Chakraborty, S. | | 登録日 | 2010-01-26 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Structural comparisons of apo- and metalated three-stranded coiled coils clarify metal binding determinants in thiolate containing designed peptides.
J.Am.Chem.Soc., 132, 2010
|
|
3TGR
 
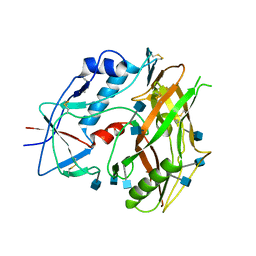 | |
3TGT
 
 | | Crystal structure of unliganded HIV-1 clade A/E strain 93TH057 gp120 core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 clade A/E 93TH057 gp120 | | 著者 | Kwon, Y.D, Kwong, P.D. | | 登録日 | 2011-08-17 | | 公開日 | 2012-04-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TGQ
 
 | |
3TGS
 
 | | Crystal structure of HIV-1 clade C strain C1086 gp120 core in complex with NBD-556 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 core, N-(4-chlorophenyl)-N'-(2,2,6,6-tetramethylpiperidin-4-yl)ethanediamide | | 著者 | Kwon, Y.D, Kwong, P.D. | | 登録日 | 2011-08-17 | | 公開日 | 2012-04-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
