7ZJ5
 
 | | Unbound state of a brocolli-pepper aptamer FRET tile. | | 分子名称: | POTASSIUM ION, brocolli-pepper aptamer | | 著者 | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | 登録日 | 2022-04-08 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.55 Å) | | 主引用文献 | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
7ZJ4
 
 | | Ligand bound state of a brocolli-pepper aptamer FRET tile | | 分子名称: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, POTASSIUM ION, ... | | 著者 | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | 登録日 | 2022-04-08 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.43 Å) | | 主引用文献 | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
7KWO
 
 | | rFVIIIFc-VWF-XTEN (BIVV001) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | 著者 | Fuller, J.R, Batchelor, J.D. | | 登録日 | 2020-12-01 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular determinants of the factor VIII/von Willebrand factor complex revealed by BIVV001 cryo-electron microscopy.
Blood, 137, 2021
|
|
7LM4
 
 | | The crystal structure of the I38T mutant PA Endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | 分子名称: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | 著者 | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | 登録日 | 2021-02-05 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
8OVR
 
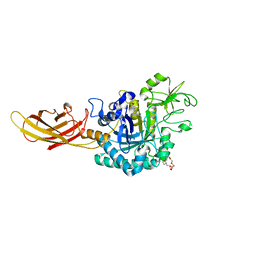 | |
8OWF
 
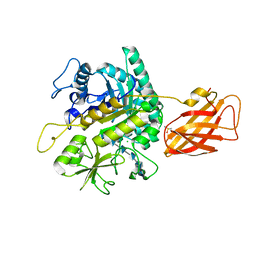 | | Clostridium perfringens chitinase CP4_3455 with chitosan | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Chitodextrinase, ... | | 著者 | Bloch, Y, Savvides, S.N. | | 登録日 | 2023-04-27 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Clostridium perfringens chitinase CP4_3455 with chitosan
To Be Published
|
|
8OXU
 
 | | Crystal Structure of the Hsp90-LA1011 Complex | | 分子名称: | ATP-dependent molecular chaperone HSP82, dimethyl 2,6-bis[2-(dimethylamino)ethyl]-1-methyl-4-[4-(trifluoromethyl)phenyl]-4~{H}-pyridine-3,5-dicarboxylate | | 著者 | Roe, S.M, Prodromou, C. | | 登録日 | 2023-05-02 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer's Disease.
Biomolecules, 13, 2023
|
|
8OOR
 
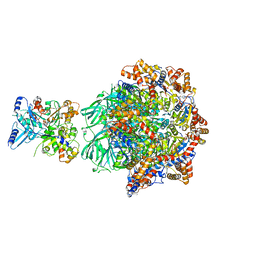 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOK
 
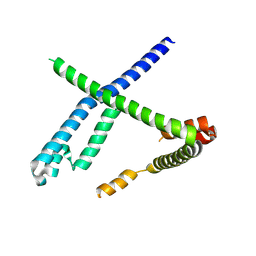 | |
8OOF
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OO7
 
 | | CryoEM Structure INO80core Hexasome complex composite model state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OO9
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-DNA refinement state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase INO80, DNA strand 1, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOT
 
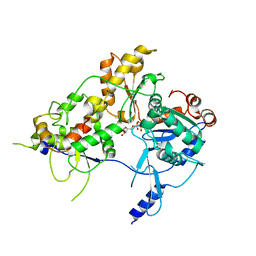 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state2 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOA
 
 | | CryoEM Structure INO80core Hexasome complex Hexasome refinement state1 | | 分子名称: | DNA Strand 2, DNA strand 1, Histone H2A, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOP
 
 | | CryoEM Structure INO80core Hexasome complex composite model state2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
7JIZ
 
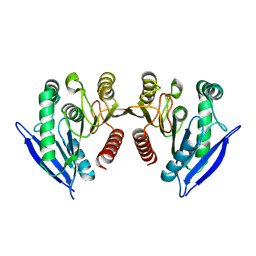 | |
7JMC
 
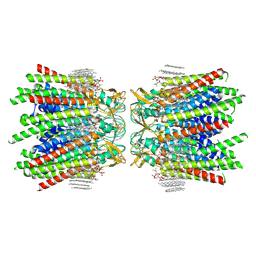 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | 著者 | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | 登録日 | 2020-07-31 | | 公開日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JY2
 
 | | Z-DNA joint X-ray/Neutron | | 分子名称: | Chains: A,B | | 著者 | Harp, J.M, Coates, L, Egli, M. | | 登録日 | 2020-08-28 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | 主引用文献 | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
7VYO
 
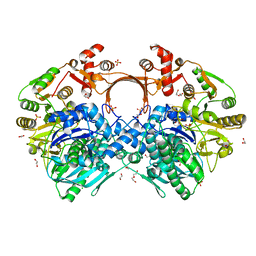 | | The structure of GdmN | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | 登録日 | 2021-11-14 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7ZGE
 
 | |
7ZF0
 
 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP and p-hydroxymandelonitrile | | 分子名称: | (2S)-HYDROXY(4-HYDROXYPHENYL)ETHANENITRILE, 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, ... | | 著者 | Putkaradze, N, Fredslund, F, Welner, D.H. | | 登録日 | 2022-03-31 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-21 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-21 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
