8OYV
 
 | |
8OYX
 
 | |
8OYW
 
 | |
8OYY
 
 | |
6CZH
 
 | | Structure of a redesigned beta barrel, mFAP0, bound to DFHBI | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP0 | | 著者 | Doyle, L.A, Stoddard, B.L. | | 登録日 | 2018-04-09 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6CZI
 
 | | Structure of a redesigned beta barrel, mFAP1, bound to DFHBI | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP1 | | 著者 | Doyle, L.A, Stoddard, B.L. | | 登録日 | 2018-04-09 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
8P49
 
 | |
8Q70
 
 | |
8QHP
 
 | |
7M0Q
 
 | |
7M5T
 
 | |
7UR7
 
 | |
7UR8
 
 | |
7UWY
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 29_bp_sh3 | | 分子名称: | De novo designed small beta-barrel protein 29_bp_sh3 | | 著者 | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | 登録日 | 2022-05-04 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UWZ
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 33_bp_sh3 | | 分子名称: | De novo designed small beta-barrel protein 33_bp_sh3 | | 著者 | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | 登録日 | 2022-05-04 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6UF2
 
 | |
6VK3
 
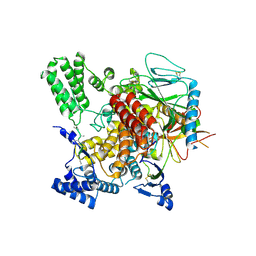 | | CryoEM structure of Hrd3/Yos9 complex | | 分子名称: | Hrd3, Protein OS-9 homolog | | 著者 | Wu, X, Rapoport, T.A. | | 登録日 | 2020-01-18 | | 公開日 | 2020-04-29 | | 最終更新日 | 2020-05-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
6VJZ
 
 | | CryoEM structure of Hrd1-Usa1/Der1/Hrd3 complex of the expected topology | | 分子名称: | Degradation in the endoplasmic reticulum protein 1, ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3, ... | | 著者 | Wu, X, Rapoport, T.A. | | 登録日 | 2020-01-18 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
6VK1
 
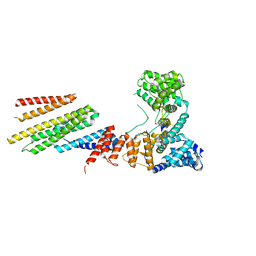 | |
6VK0
 
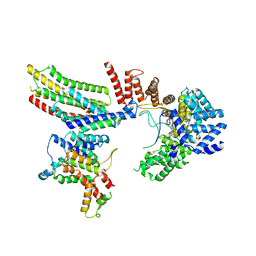 | | CryoEM structure of Hrd1-Usa1/Der1/Hrd3 of the flipped topology | | 分子名称: | Degradation in the endoplasmic reticulum protein 1, ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3, ... | | 著者 | Wu, X, Rapoport, T.A. | | 登録日 | 2020-01-18 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
