5HJ7
 
 | | Glutamate Racemase Mycobacterium tuberculosis (MurI) with bound D-glutamate, 2.3 Angstrom resolution, X-ray diffraction | | 分子名称: | D-GLUTAMIC ACID, Glutamate racemase | | 著者 | Poen, S, Nakatani, Y, Krause, K. | | 登録日 | 2016-01-12 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Exploring the structure of glutamate racemase from Mycobacterium tuberculosis as a template for anti-mycobacterial drug discovery.
Biochem. J., 473, 2016
|
|
5IJW
 
 | | Glutamate Racemase (MurI) from Mycobacterium smegmatis with bound D-glutamate, 1.8 Angstrom resolution, X-ray diffraction | | 分子名称: | D-GLUTAMIC ACID, Glutamate racemase, IODIDE ION | | 著者 | Poen, S, Nakatani, Y, Krause, K. | | 登録日 | 2016-03-02 | | 公開日 | 2016-05-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Exploring the structure of glutamate racemase from Mycobacterium tuberculosis as a template for anti-mycobacterial drug discovery.
Biochem. J., 473, 2016
|
|
5KMQ
 
 | |
4QHR
 
 | | The structure of alanine racemase from Acinetobacter baumannii | | 分子名称: | Alanine racemase | | 著者 | Davis, E, Scaletti-Hutchinson, E, Nakatani, Y, Krause, K.L. | | 登録日 | 2014-05-29 | | 公開日 | 2015-05-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structure of alanine racemase from Acinetobacter baumannii
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
4OYQ
 
 | | (6-isothiocyanatohexyl)benzene inhibitor complexed with Macrophage Migration Inhibitory Factor | | 分子名称: | 6-isothiocyanatohexylbenzene, GLYCEROL, Macrophage migration inhibitory factor, ... | | 著者 | Spencer, E.S, Dale, E.J, Gommans, A.L, Vo, C.T, Rutledge, M.T, Nakatani, Y, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | 登録日 | 2014-02-12 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | To be published
To be published
|
|
4OSF
 
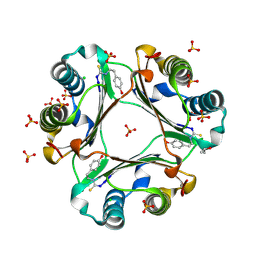 | | 4-(2-isothiocyanatoethyl)phenol inhibitor complexed with Macrophage Migration Inhibitory Factor | | 分子名称: | CHLORIDE ION, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | 著者 | Spencer, E.S, Dale, E.J, Gommans, A.L, Vo, C.T, Rutledge, M.T, Nakatani, Y, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | 登録日 | 2014-02-12 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
1TAF
 
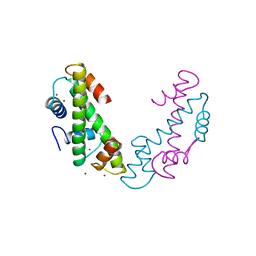 | | DROSOPHILA TBP ASSOCIATED FACTORS DTAFII42/DTAFII62 HETEROTETRAMER | | 分子名称: | TFIID TBP ASSOCIATED FACTOR 42, TFIID TBP ASSOCIATED FACTOR 62, ZINC ION | | 著者 | Xie, X, Kokubo, T, Cohen, S.L, Mirza, U.A, Hoffmann, A, Chait, B.T, Roeder, R.G, Nakatani, Y, Burley, S.K. | | 登録日 | 1996-06-01 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural similarity between TAFs and the heterotetrameric core of the histone octamer.
Nature, 380, 1996
|
|
1TBA
 
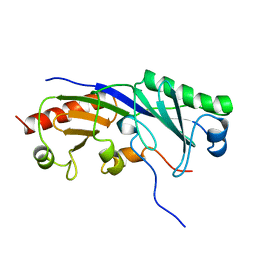 | | SOLUTION STRUCTURE OF A TBP-TAFII230 COMPLEX: PROTEIN MIMICRY OF THE MINOR GROOVE SURFACE OF THE TATA BOX UNWOUND BY TBP, NMR, 25 STRUCTURES | | 分子名称: | TRANSCRIPTION INITIATION FACTOR IID 230K CHAIN, TRANSCRIPTION INITIATION FACTOR TFIID | | 著者 | Liu, D, Ishima, R, Tong, K.I, Bagby, S, Kokubo, T, Muhandiram, D.R, Kay, L.E, Nakatani, Y, Ikura, M. | | 登録日 | 1998-08-16 | | 公開日 | 1999-08-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP.
Cell(Cambridge,Mass.), 94, 1998
|
|
4KQ6
 
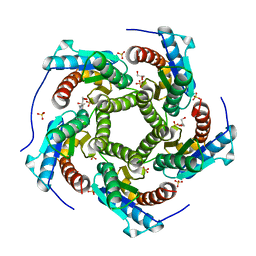 | | Product complex of lumazine synthase from candida glabrata | | 分子名称: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, 6,7-dimethyl-8-ribityllumazine synthase, GLYCEROL, ... | | 著者 | Shankar, M, Wilbanks, S.M, Nakatani, Y, Monk, B.C, Tyndall, J.D.A. | | 登録日 | 2013-05-14 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Catalysis product captured in lumazine synthase from the fungal pathogen Candida glabrata.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4ZK9
 
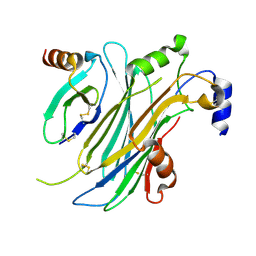 | | The chemokine binding protein of orf virus complexed with CCL2 | | 分子名称: | C-C motif chemokine 2, Chemokine binding protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Knapp, K.M, Nakatani, Y, Krause, K.L. | | 登録日 | 2015-04-30 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of Orf Virus Chemokine Binding Protein in Complex with Host Chemokines Reveal Clues to Broad Binding Specificity.
Structure, 23, 2015
|
|
4ZKB
 
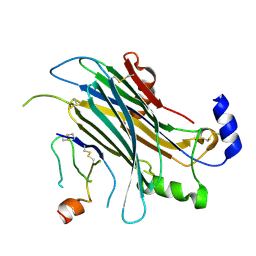 | | The chemokine binding protein of orf virus complexed with CCL3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 3, Chemokine binding protein, ... | | 著者 | Knapp, K.M, Nakatani, Y, Krause, K.L. | | 登録日 | 2015-04-30 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of Orf Virus Chemokine Binding Protein in Complex with Host Chemokines Reveal Clues to Broad Binding Specificity.
Structure, 23, 2015
|
|
5CEK
 
 | |
5CEM
 
 | |
2E8I
 
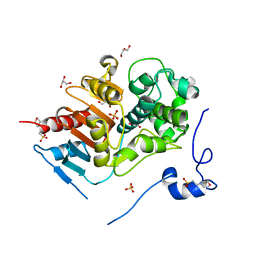 | | Structure of 6-aminohexanoate-dimer hydrolase, D1 mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Shibata, N, Higuchi, Y, Negoro, S. | | 登録日 | 2007-01-20 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
2ZM0
 
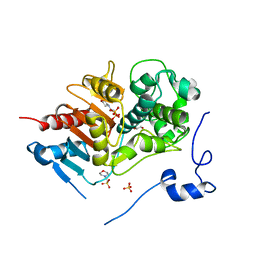 | | Structure of 6-aminohexanoate-dimer hydrolase, G181D/H266N/D370Y mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | 登録日 | 2008-04-10 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
2ZMA
 
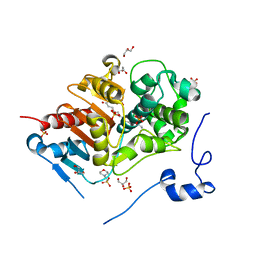 | | Crystal Structure of 6-Aminohexanoate-dimer Hydrolase S112A/G181D/H266N/D370Y Mutant with Substrate | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | 登録日 | 2008-04-14 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
2ZM7
 
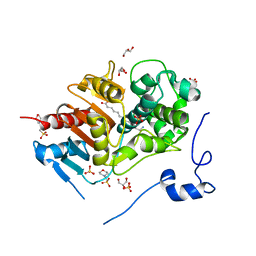 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/G181D Mutant Complexed with 6-Aminohexanoate-dimer | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | 登録日 | 2008-04-14 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
3EI2
 
 | |
3EI4
 
 | |
3EI3
 
 | | Structure of the hsDDB1-drDDB2 complex | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | 著者 | Scrima, A, Thoma, N.H. | | 登録日 | 2008-09-15 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EI1
 
 | |
2PC8
 
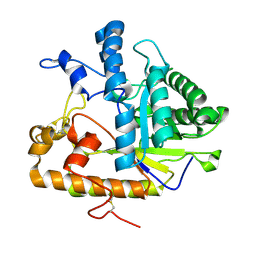 | | E292Q mutant of EXO-B-(1,3)-Glucanase from Candida Albicans in complex with two separately bound glucopyranoside units at 1.8 A | | 分子名称: | Hypothetical protein XOG1, beta-D-glucopyranose | | 著者 | Cutfield, S.M, Cutfield, J.F, Patrick, W.M. | | 登録日 | 2007-03-29 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Carbohydrate binding sites in Candida albicans exo-beta-1,3-glucanase and the role of the Phe-Phe 'clamp' at the active site entrance.
Febs J., 277, 2010
|
|
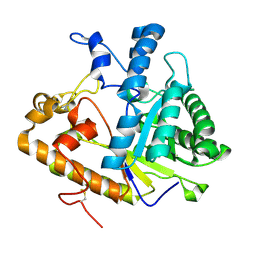
2PF0
 
 | |
4P5I
 
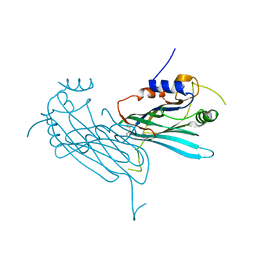 | | Crystal structure of the chemokine binding protein from orf virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chemokine binding protein | | 著者 | Counago, R.M, Krause, K.L. | | 登録日 | 2014-03-17 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structures of Orf Virus Chemokine Binding Protein in Complex with Host Chemokines Reveal Clues to Broad Binding Specificity.
Structure, 23, 2015
|
|
3WNR
 
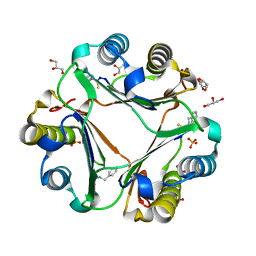 | | Multiple binding modes of benzyl isothiocyanate inhibitor complexed with Macrophage Migration Inhibitory Factor | | 分子名称: | CHLORIDE ION, GLYCEROL, Macrophage migration inhibitory factor, ... | | 著者 | Spencer, E.S, Dale, E.J, Gommans, A.L, Rutledge, M.T, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | 登録日 | 2013-12-16 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.008 Å) | | 主引用文献 | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
