7EXB
 
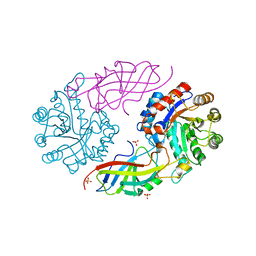 | | DfgA-DfgB complex apo 2.4 angstrom | | 分子名称: | DfgB, MANGANESE (II) ION, SULFATE ION, ... | | 著者 | Mori, T, Senda, M, Senda, T, Abe, I. | | 登録日 | 2021-05-26 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
6M35
 
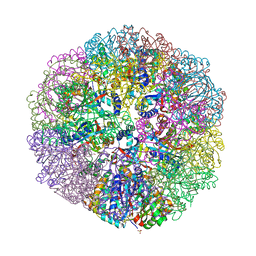 | | Crystal structure of sulfur oxygenase reductase from Sulfurisphaera tokodaii | | 分子名称: | FE (III) ION, GLYCEROL, SULFATE ION, ... | | 著者 | Sato, Y, Yabuki, T, Arakawa, T, Yamada, C, Fushinobu, S, Wakagi, T. | | 登録日 | 2020-03-02 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase fromSulfurisphaera tokodaii.
J Struct Biol X, 4, 2020
|
|
6LTB
 
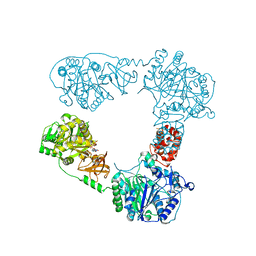 | |
6LTA
 
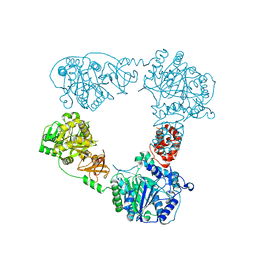 | |
6LTC
 
 | |
6LTD
 
 | |
7BVR
 
 | | DgpB-DgpC complex apo | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | 著者 | Mori, T, He, H, Abe, I. | | 登録日 | 2020-04-11 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7BVS
 
 | | DfgA-DfgB complex apo | | 分子名称: | DfgB, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Mori, T, He, H, Abe, I. | | 登録日 | 2020-04-11 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7EFV
 
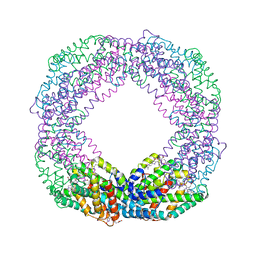 | | Crystal structure of octameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | 分子名称: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | 著者 | Minato, T, Teramoto, T, Hung, N.K, Yamada, K, Ogo, S, Kakuta, Y, Yoon, K.S. | | 登録日 | 2021-03-23 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7EFW
 
 | | Crystal structure of hexameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | 分子名称: | C-phycocyanin alpha chain, C-phycocyanin beta chain, GLYCEROL, ... | | 著者 | Minato, T, Teramoto, T, Hung, N.K, Yamada, K, Ogo, S, Kakuta, Y, Yoon, K.S. | | 登録日 | 2021-03-23 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7VTA
 
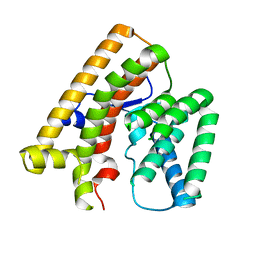 | |
7VTB
 
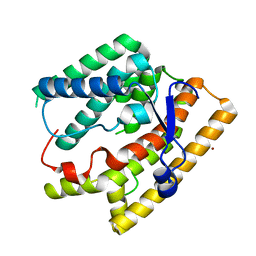 | |
7WIJ
 
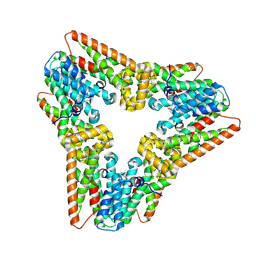 | |
7VU9
 
 | |
7WJE
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerotetraose | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJF
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with kojibiose | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-2)-alpha-D-glucopyranose | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJD
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerotriose | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, ... | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJ9
 
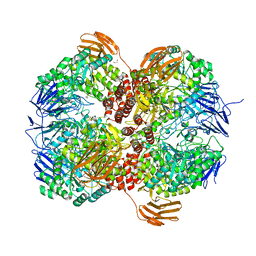 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase, P21 space group | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, Xylitol | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJB
 
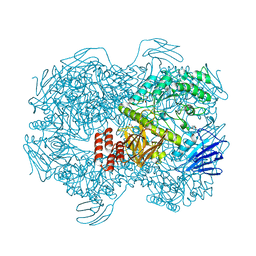 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase in complex with glucose | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose, ... | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJC
 
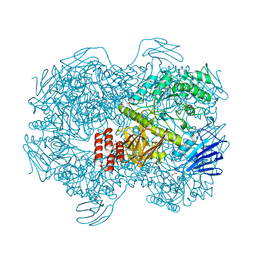 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerose | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJA
 
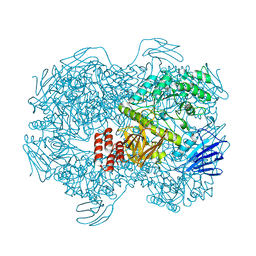 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase, P6322 space group | | 分子名称: | 1,2-ETHANEDIOL, Alpha-xylosidase | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7B67
 
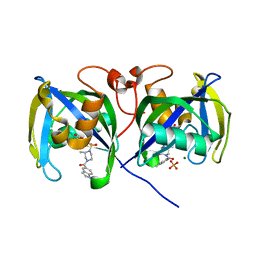 | | Structure of NUDT15 V18_V19insGV Mutant in complex with TH7755 | | 分子名称: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15, ... | | 著者 | Rehling, D, Stenmark, P. | | 登録日 | 2020-12-07 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B65
 
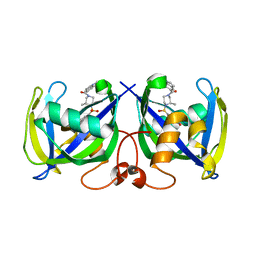 | | Structure of NUDT15 R139C Mutant in complex with TH7755 | | 分子名称: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | 著者 | Rehling, D, Stenmark, P. | | 登録日 | 2020-12-07 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B63
 
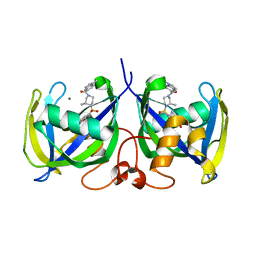 | | Structure of NUDT15 in complex with TH7755 | | 分子名称: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, MAGNESIUM ION, Probable 8-oxo-dGTP diphosphatase NUDT15 | | 著者 | Rehling, D, Stenmark, P. | | 登録日 | 2020-12-07 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B64
 
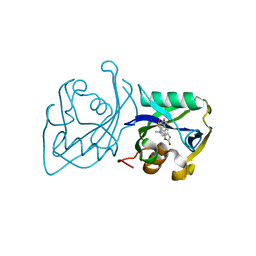 | | Structure of NUDT15 V18I Mutant in complex with TH7755 | | 分子名称: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | 著者 | Rehling, D, Stenmark, P. | | 登録日 | 2020-12-07 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
