8XEY
 
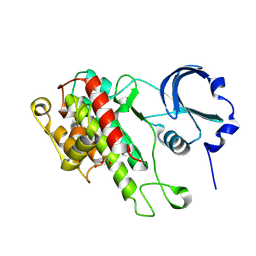 | | The Crystal Structure of C-terminal kinase domain of RSK2 from Biortus | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ribosomal protein S6 kinase alpha-3 | | 著者 | Wang, F, Cheng, W, Lv, Z, Meng, Q, Zhang, B. | | 登録日 | 2023-12-13 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | The Crystal Structure of C-terminal kinase domain of RSK2 from Biortus
To Be Published
|
|
6TB0
 
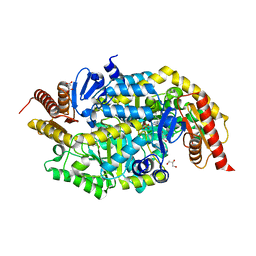 | | Crystal structure of thermostable omega transaminase 4-fold mutant from Pseudomonas jessenii | | 分子名称: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | 登録日 | 2019-10-31 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
6TB1
 
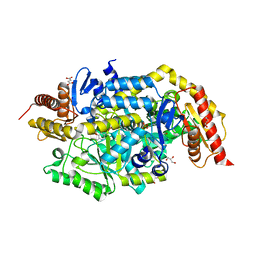 | | Crystal structure of thermostable omega transaminase 6-fold mutant from Pseudomonas jessenii | | 分子名称: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | 登録日 | 2019-10-31 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
6OOD
 
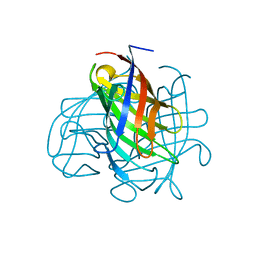 | | Structure of the pterocarpan synthase dirigent protein PsPTS1 | | 分子名称: | pterocarpan synthase dirigent protein PsPTS1 | | 著者 | Smith, C.A. | | 登録日 | 2019-04-23 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Pterocarpan synthase (PTS) structures suggest a common quinone methide-stabilizing function in dirigent proteins and proteins with dirigent-like domains.
J.Biol.Chem., 295, 2020
|
|
6OOC
 
 | |
7B4J
 
 | | Thermostable omega transaminase PjTA-R6 variant W58M/F86L/R417L engineered for asymmetric synthesis of enantiopure bulky amines | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase family protein, SUCCINIC ACID | | 著者 | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | 登録日 | 2020-12-02 | | 公開日 | 2021-09-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational Redesign of an omega-Transaminase from Pseudomonas jessenii for Asymmetric Synthesis of Enantiopure Bulky Amines.
Acs Catalysis, 11, 2021
|
|
7B4I
 
 | | Thermostable omega transaminase PjTA-R6 variant W58G engineered for asymmetric synthesis of enantiopure bulky amines | | 分子名称: | Aspartate aminotransferase family protein, PYRIDOXAL-5'-PHOSPHATE, SUCCINIC ACID | | 著者 | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | 登録日 | 2020-12-02 | | 公開日 | 2021-09-01 | | 最終更新日 | 2021-09-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Computational Redesign of an omega-Transaminase from Pseudomonas jessenii for Asymmetric Synthesis of Enantiopure Bulky Amines.
Acs Catalysis, 11, 2021
|
|
4BMC
 
 | | Crystal structure of s.pombe Rad4 BRCT1,2 | | 分子名称: | CHLORIDE ION, S-M CHECKPOINT CONTROL PROTEIN RAD4 | | 著者 | Meng, Q, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | 登録日 | 2013-05-07 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.982 Å) | | 主引用文献 | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
4BMD
 
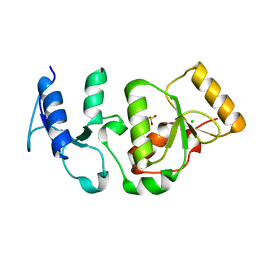 | | Crystal structure of S.pombe Rad4 BRCT3,4 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, S-M CHECKPOINT CONTROL PROTEIN RAD4 | | 著者 | Meng, Q, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | 登録日 | 2013-05-07 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
1JE9
 
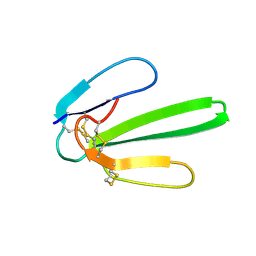 | | NMR SOLUTION STRUCTURE OF NT2 | | 分子名称: | SHORT NEUROTOXIN II | | 著者 | Cheng, Y, Wang, W, Wang, J. | | 登録日 | 2001-06-16 | | 公開日 | 2001-07-04 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function relationship of three neurotoxins from the venom of Naja kaouthia: a comparison between the NMR-derived structure of NT2 with its homologues, NT1 and NT3
BIOCHIM.BIOPHYS.ACTA, 1594, 2002
|
|
7YP0
 
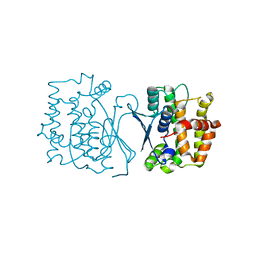 | | Crystal structure of CtGST | | 分子名称: | Glutathione S-transferase | | 著者 | Yang, J, Fan, J.P, Lei, X.G. | | 登録日 | 2022-08-02 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7YOC
 
 | |
6G4C
 
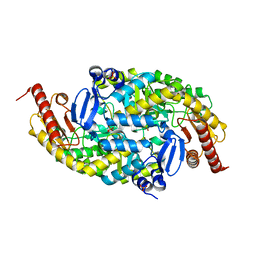 | |
6G4F
 
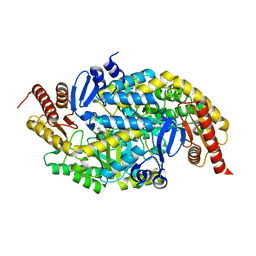 | |
6G4B
 
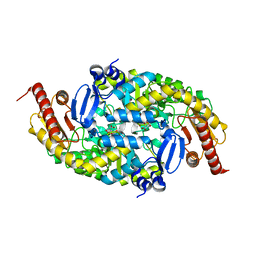 | |
6G4D
 
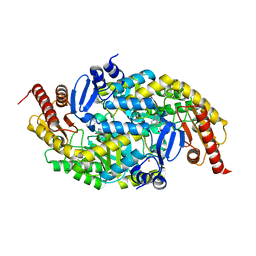 | |
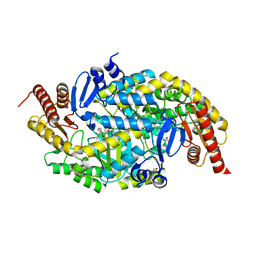
6G4E
 
 | |
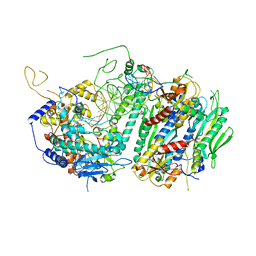
4ZTZ
 
 | |
2LTH
 
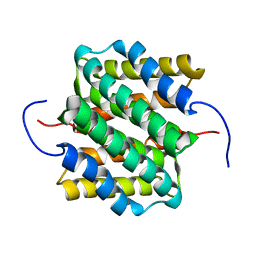 | | NMR structure of major ampullate spidroin 1 N-terminal domain at pH 5.5 | | 分子名称: | Major ampullate spidroin 1 | | 著者 | Otikovs, M, Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S, Johansson, J. | | 登録日 | 2012-05-25 | | 公開日 | 2013-11-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation.
Nat Commun, 5, 2014
|
|
2MX9
 
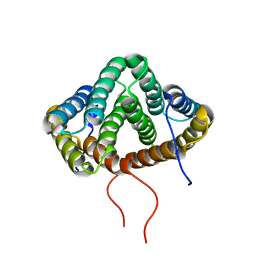 | | NMR structure of N-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) at pH 5.5 | | 分子名称: | Minor ampullate spidroin | | 著者 | Otikovs, M, Jaudzems, K, Chen, G, Nordling, K, Rising, A, Johansson, J. | | 登録日 | 2014-12-17 | | 公開日 | 2015-08-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Diversified Structural Basis of a Conserved Molecular Mechanism for pH-Dependent Dimerization in Spider Silk N-Terminal Domains.
Chembiochem, 16, 2015
|
|
2MX8
 
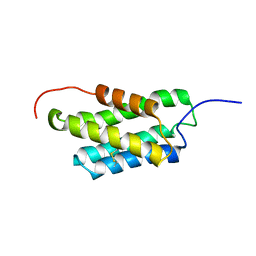 | | NMR structure of N-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) at pH 7.2 | | 分子名称: | Minor ampullate spidroin | | 著者 | Otikovs, M, Jaudzems, K, Chen, G, Nordling, K, Rising, A, Johansson, J. | | 登録日 | 2014-12-17 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Diversified Structural Basis of a Conserved Molecular Mechanism for pH-Dependent Dimerization in Spider Silk N-Terminal Domains.
Chembiochem, 16, 2015
|
|
6A83
 
 | |
6A82
 
 | |
8K2O
 
 | |
8K2P
 
 | |
